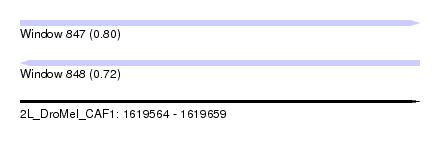
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,619,564 – 1,619,659 |
| Length | 95 |
| Max. P | 0.795185 |

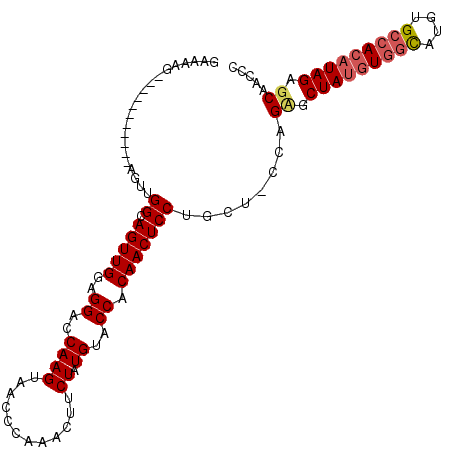
| Location | 1,619,564 – 1,619,659 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
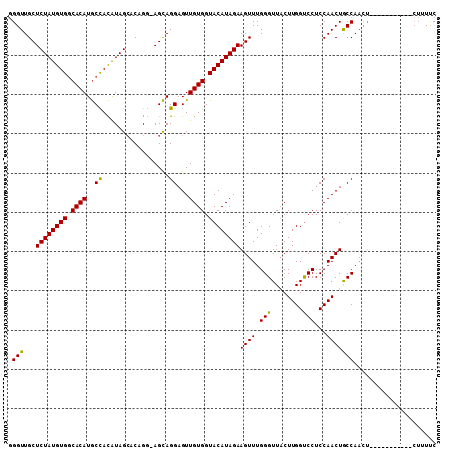
>2L_DroMel_CAF1 1619564 95 + 22407834 CGGUUGCUCUAUGUGGCACAUACUAUGUAGCGCUGCGAGUAGGAGUUGUGGUACAUAGAAGUUUGGGUUCCUUGGUCCUCCAACUGCCAACU--------------UCC .((((((((((((((.((((..((((...((...))..))))....)))).))))))))((((.(((..(....)..))).))))).)))))--------------... ( -29.40) >DroEre_CAF1 99686 94 + 1 GGGUUGCUCUAUGUGGCACAUGCCACAUAGAACGGG-AGCAGGAGUUGUGGUACAUAGAAGUUUGGGUUACUUGGUCCUCCAACUACCA--------------CUUUUC ..(((.(((((((((((....))))))))))...).-)))..(((..((((((((........))......((((....)))).)))))--------------)..))) ( -29.50) >DroYak_CAF1 98558 109 + 1 GGGUUGCUCUAUGUGCCACAUGCCACAUAGCUCAGGCAGCAGGAGUUGUGGUACAUAGAAGUUUGGGUUACUUGGCCCUCCAACUGCCAACUGCCUACACCCACUUUUC ((((.(((((((((((((((..((.....((....))....))...)))))))))))).))).(((((...(((((.........)))))..))))).))))....... ( -39.80) >consensus GGGUUGCUCUAUGUGGCACAUGCCACAUAGCACAGG_AGCAGGAGUUGUGGUACAUAGAAGUUUGGGUUACUUGGUCCUCCAACUGCCAACU___________CUUUUC .(((...((((((((.((((..((.................))...)))).))))))))((((.(((........)))...)))))))..................... (-21.89 = -21.23 + -0.66)

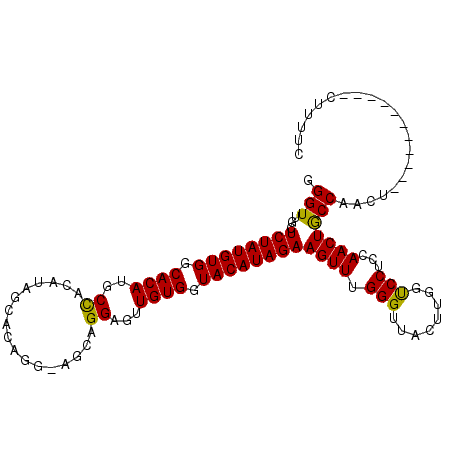
| Location | 1,619,564 – 1,619,659 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -32.19 |
| Consensus MFE | -17.66 |
| Energy contribution | -19.00 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1619564 95 - 22407834 GGA--------------AGUUGGCAGUUGGAGGACCAAGGAACCCAAACUUCUAUGUACCACAACUCCUACUCGCAGCGCUACAUAGUAUGUGCCACAUAGAGCAACCG ((.--------------(((.((.(((((..((..((.((((.......)))).))..)).))))))).)))....(((((((...))).))))............)). ( -20.50) >DroEre_CAF1 99686 94 - 1 GAAAAG--------------UGGUAGUUGGAGGACCAAGUAACCCAAACUUCUAUGUACCACAACUCCUGCU-CCCGUUCUAUGUGGCAUGUGCCACAUAGAGCAACCC .....(--------------(((((..((((((...............))))))..))))))..........-...((((((((((((....))))))))))))..... ( -34.66) >DroYak_CAF1 98558 109 - 1 GAAAAGUGGGUGUAGGCAGUUGGCAGUUGGAGGGCCAAGUAACCCAAACUUCUAUGUACCACAACUCCUGCUGCCUGAGCUAUGUGGCAUGUGGCACAUAGAGCAACCC .......((((..(((((((.((.(((((..(((........)))..((......))....))))))).)))))))(..(((((((.(....).)))))))..).)))) ( -41.40) >consensus GAAAAG___________AGUUGGCAGUUGGAGGACCAAGUAACCCAAACUUCUAUGUACCACAACUCCUGCU_CCAGAGCUAUGUGGCAUGUGCCACAUAGAGCAACCC .....................((.(((((..((..((((............)).))..)).)))))))........((.(((((((((....))))))))).))..... (-17.66 = -19.00 + 1.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:27 2006