| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,897,580 – 14,897,768 |
| Length | 188 |
| Max. P | 0.870306 |

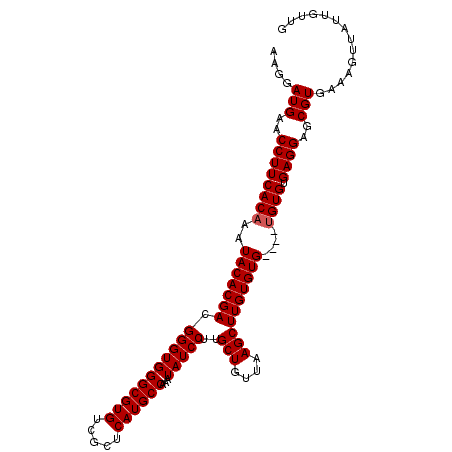
| Location | 14,897,580 – 14,897,690 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -13.30 |
| Energy contribution | -13.30 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
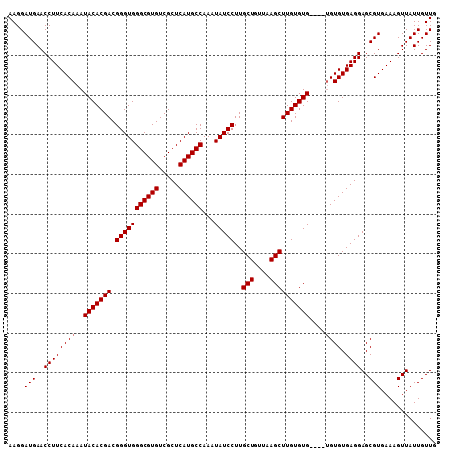
>2L_DroMel_CAF1 14897580 110 + 22407834 CACCCGCUCCUGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUCACGCUCCUCACACACA ...........(((((...........((....))((((((((((.....................))))))))))....................)))))......... ( -18.40) >DroPse_CAF1 115508 106 + 1 GUGGCUCGGUCGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUU----CCCUCACACACA (((((((.....))))..............((((.((((((((((.....................))))))))))....))))..........----......)))... ( -19.50) >DroSec_CAF1 55259 108 + 1 CACCCUCUCCUGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUCACGCUCCUCACACU-- ...........(((((...........((....))((((((((((.....................))))))))))....................))))).......-- ( -18.40) >DroSim_CAF1 55135 108 + 1 CACCCUCUCCUGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUCACGCUCCUCACACA-- ...........(((((...........((....))((((((((((.....................))))))))))....................))))).......-- ( -18.40) >DroPer_CAF1 98085 106 + 1 GUGGCUCGGUCGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUU----CCCUCACACACA (((((((.....))))..............((((.((((((((((.....................))))))))))....))))..........----......)))... ( -19.50) >consensus CACCCUCUCCUGGAGCAUAUCUAUUCUGCAGUUGCUGUCAACAUUUUUCAUAUCAUAAACACUUUUAGUGUUGAUAUUACCAACAAUAACUUUCACGCUCCUCACACACA ...........((.(((.........))).((((.((((((((((.....................))))))))))....))))...............))......... (-13.30 = -13.30 + -0.00)

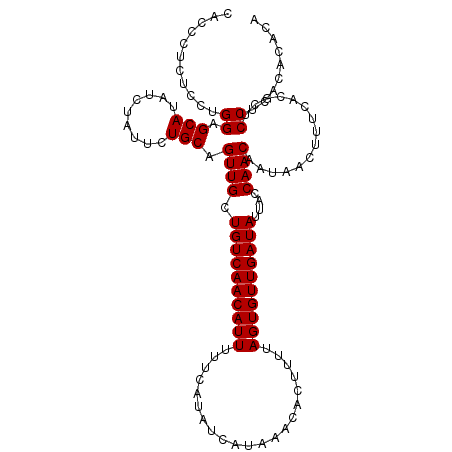

| Location | 14,897,580 – 14,897,690 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.02 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14897580 110 - 22407834 UGUGUGUGAGGAGCGUGAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCAGGAGCGGGUG ..(((....((((((((...((((..(((....((((((((((.....))))...))))))....)))..)))).((....))........))))))))....))).... ( -26.70) >DroPse_CAF1 115508 106 - 1 UGUGUGUGAGGG----AAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCGACCGAGCCAC .(((.((..((.----...((((.(((((..((((((((((((.....))))...))))))......))..))))).))))(((.........))).....))..))))) ( -21.60) >DroSec_CAF1 55259 108 - 1 --AGUGUGAGGAGCGUGAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCAGGAGAGGGUG --.......((((((((...((((..(((....((((((((((.....))))...))))))....)))..)))).((....))........))))))))........... ( -26.10) >DroSim_CAF1 55135 108 - 1 --UGUGUGAGGAGCGUGAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCAGGAGAGGGUG --.......((((((((...((((..(((....((((((((((.....))))...))))))....)))..)))).((....))........))))))))........... ( -26.10) >DroPer_CAF1 98085 106 - 1 UGUGUGUGAGGG----AAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCGACCGAGCCAC .(((.((..((.----...((((.(((((..((((((((((((.....))))...))))))......))..))))).))))(((.........))).....))..))))) ( -21.60) >consensus UGUGUGUGAGGAGCGUGAAAGUUAUUGUUGGUAAUAUCAACACUAAAAGUGUUUAUGAUAUGAAAAAUGUUGACAGCAACUGCAGAAUAGAUAUGCUCCAGGAGAGGGUG .........((((((((...((((..(((....((((((((((.....))))...))))))....)))..)))).((....))........))))))))........... (-20.90 = -22.02 + 1.12)

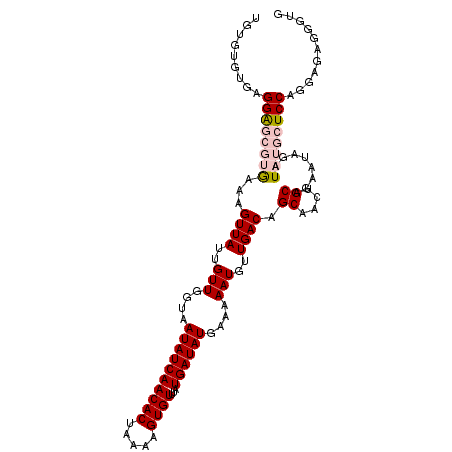
| Location | 14,897,660 – 14,897,768 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 96.88 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -30.37 |
| Energy contribution | -30.70 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14897660 108 - 22407834 AAGGAUGAACCUUCACAAAUACACGACGGGUGGGCGUGUCGCUCAUGCCAAAUAUCCUUGCUGUUAAGCUUGUGUGUGUGUGUGUGAGGAGCGUGAAAGUUAUUGUUG ....(((..((((((((.((((((((.(((((((((((.....))))))...)))))..(((....)))))))))))...)))).))))..))).............. ( -34.30) >DroSec_CAF1 55339 104 - 1 AAGGAUGAACCUUCACAAAUACACGACGGGUGGGCGUGUCGCUCAUGCCAAAUAUCCUUGCUGUUAAGCUUGUGUG----AGUGUGAGGAGCGUGAAAGUUAUUGUUG ...((..(..(((((((..(((((((.(((((((((((.....))))))...)))))..(((....))))))))))----..)))))))(((......))).)..)). ( -33.00) >DroSim_CAF1 55215 104 - 1 AAGGAUGAACCUUCACAAAUACACGACGGGUGGGCGUGUCGCUCAUGCCAAAUAUCCUUGCUGUUAAGCUUGUGUG----UGUGUGAGGAGCGUGAAAGUUAUUGUUG ...((..(..(((((((.((((((((.(((((((((((.....))))))...)))))..(((....))))))))))----).)))))))(((......))).)..)). ( -34.20) >consensus AAGGAUGAACCUUCACAAAUACACGACGGGUGGGCGUGUCGCUCAUGCCAAAUAUCCUUGCUGUUAAGCUUGUGUG____UGUGUGAGGAGCGUGAAAGUUAUUGUUG ....(((..((((((((..(((((((.(((((((((((.....))))))...)))))..(((....))))))))))....)))).))))..))).............. (-30.37 = -30.70 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:31 2006