
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,897,108 – 14,897,211 |
| Length | 103 |
| Max. P | 0.500000 |

| Location | 14,897,108 – 14,897,211 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -22.75 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
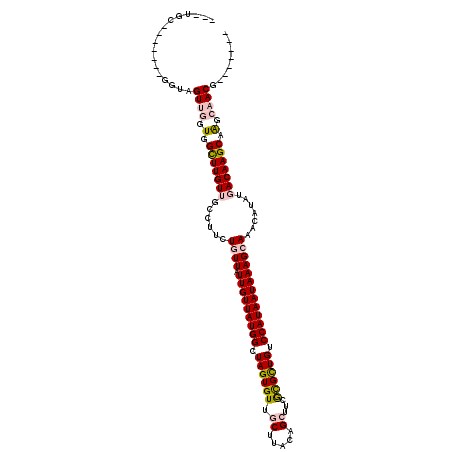
>2L_DroMel_CAF1 14897108 103 + 22407834 ---UGU-------GGUAGUUGGUGGCUUGUUGCCUUCUGUUAUUGUUAUGGCUAGUGUUGCUUACAGCUUCGCGCUGUCCAUAAUAAAGCAAACAUAUGACAAGCAAGCAACG------- ---...-------....((((.(.(((((((......((((.(((((((((.((((((.((.....))...)))))).))))))))))))).......))))))).).)))).------- ( -33.82) >DroPse_CAF1 114967 113 + 1 GUCGUC-------GGUCGUCAGUUGUUUGUAGCUCGUUCUUAUUGUUAUGGCUAGUGUUUCUUACGGCUUCAUGUUGUCCAUAAUAAAGCAAACAUAUGACAAGCCAGAAACAAGAAAGG ......-------(((.((((..(((((((((......))..(((((((((............(((((.....)))))))))))))).)))))))..))))..))).............. ( -22.80) >DroSec_CAF1 54787 103 + 1 ---UGU-------GGUAGUUGGUGGCUUGUUGCCUUCUGUUAUUGUUAUGGCUAGUGUUGCUUACAGCUUCGCGCUGUCCAUAAUAAAGCAAACAUAUGACAAGCAAGCAACG------- ---...-------....((((.(.(((((((......((((.(((((((((.((((((.((.....))...)))))).))))))))))))).......))))))).).)))).------- ( -33.82) >DroSim_CAF1 54663 103 + 1 ---UGU-------GGUAGUUGGUGGCUUGUUUCCUUCUGUUAUUGUUAUGGCUAGUGUUGCUUACAGCUUCGCGCUGUCCAUAAUAAAGCAAACAUAUGACAAGCAGGCAACG------- ---...-------....((((.(.(((((((......((((.(((((((((.((((((.((.....))...)))))).))))))))))))).......))))))).).)))).------- ( -34.52) >DroYak_CAF1 54317 103 + 1 ---AGC-------GGUAGUUGGUGGCUUGUUGCCUUCUGUUAUUGUUAUGGCUAGUGUUGCUUACAGCUUCGUGUUGUCCAUAAUAAAGCAAACAUAUGACAAGCAAGCAACG------- ---...-------....((((.(.(((((((......((((.(((((((((.(((..(.((.....))...)..))).))))))))))))).......))))))).).)))).------- ( -28.02) >DroPer_CAF1 97555 120 + 1 GUCGUCGGUCGUCGGUCGUCAGUUGUUUGUAGCUCGUUCUUAUUGUUAUGGCUAGUGUUUCUUACGGCUUCAUGUUGUCCAUAAUAAAGCAAACAUAUGACAAGCCAGAAACAAGAAAGG ....((.....(((((.((((..(((((((((......))..(((((((((............(((((.....)))))))))))))).)))))))..))))..))).)).....)).... ( -23.70) >consensus ___UGC_______GGUAGUUGGUGGCUUGUUGCCUUCUGUUAUUGUUAUGGCUAGUGUUGCUUACAGCUUCGCGCUGUCCAUAAUAAAGCAAACAUAUGACAAGCAAGCAACG_______ .................((((.(.(((((((......((((.(((((((((.((((((.((.....))...)))))).))))))))))))).......))))))).).))))........ (-22.75 = -23.34 + 0.58)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:28 2006