
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
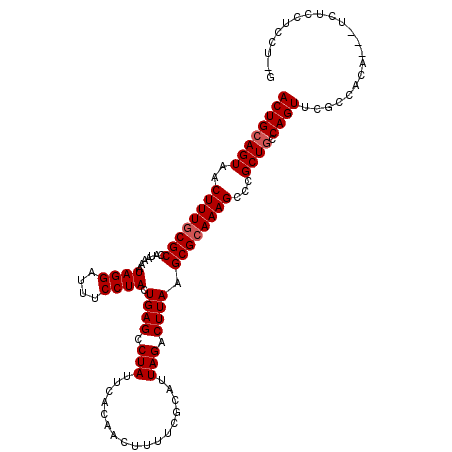
| Location | 14,890,376 – 14,890,487 |
| Length | 111 |
| Max. P | 0.997923 |

| Location | 14,890,376 – 14,890,487 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.50 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
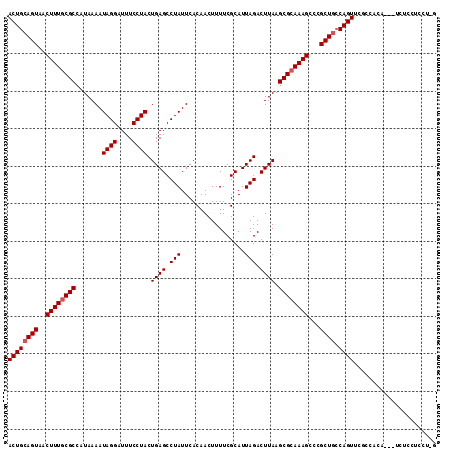
>2L_DroMel_CAF1 14890376 111 + 22407834 ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCCCGCUGCCAGUUCGCCACA---UCUCCUCUUCG ((((((((..((((((((.......((((....)))).((((.(((.................))).)))).))))))))...)))).))))........---........... ( -26.23) >DroSim_CAF1 47767 111 + 1 ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCCCGCUGCCAGUUCGCCACA---UCUCCUCCUCG ((((((((..((((((((.......((((....)))).((((.(((.................))).)))).))))))))...)))).))))........---........... ( -26.23) >DroEre_CAF1 47590 110 + 1 ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCCCGCUACCAGUUCGCCACA---UCUCCUCCA-G ((((.(((..((((((((.......((((....)))).((((.(((.................))).)))).))))))))...)))..))))........---.........-. ( -21.53) >DroYak_CAF1 47359 110 + 1 ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCCCGCUGCCAGUUCGCCACA---UCUCCUUCU-G ((((((((..((((((((.......((((....)))).((((.(((.................))).)))).))))))))...)))).))))........---.........-. ( -26.23) >DroAna_CAF1 50512 108 + 1 ACUGCAGUAACUUUACGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGAAAAGCUCGCUGCCAGUGUUCUUC-----CUCCUGCU-U ((((((((.................((((....))))..((((...........(((((((...........))))))))))))))).)))).......-----........-. ( -22.20) >DroPer_CAF1 87466 113 + 1 ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCUCGCUGGCAGUUCGCUCCUCGGUGGCCUUUC-C (((((((..(((((((((.......((((....)))).((((.(((.................))).)))).)))))))).)..)).)))))(((((....)))))......-. ( -28.83) >consensus ACUGCAGUAACUUUGCGCCAUAAAAUAGGAUUUCCUACUGAGCCUAUUCACAACUUUUCGCAUUAGACUUAAGCGCAAAGCCCGCUGCCAGUUCGCCACA___UCUCCUCCU_G ((((((((..((((((((.......((((....)))).((((.(((.................))).)))).))))))))...)))).))))...................... (-24.16 = -24.50 + 0.33)

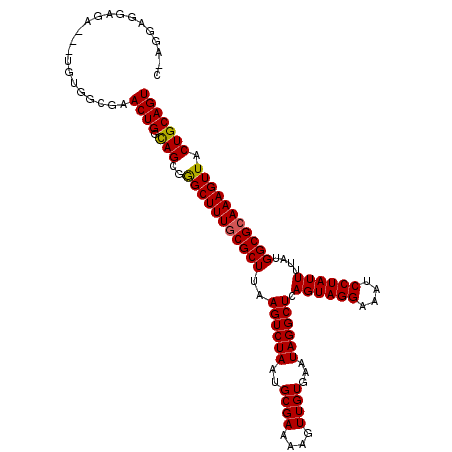
| Location | 14,890,376 – 14,890,487 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -32.78 |
| Energy contribution | -32.58 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
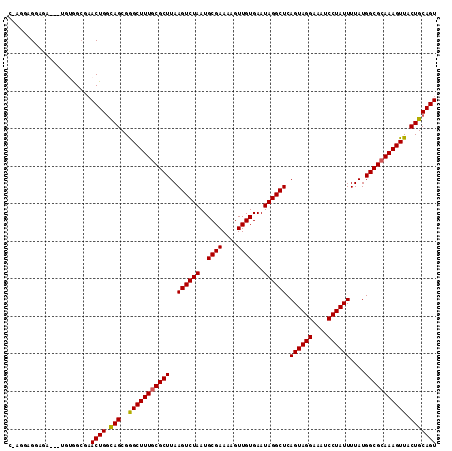
>2L_DroMel_CAF1 14890376 111 - 22407834 CGAAGAGGAGA---UGUGGCGAACUGGCAGCGGGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU ...........---........((((.(((..(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).))))))) ( -33.80) >DroSim_CAF1 47767 111 - 1 CGAGGAGGAGA---UGUGGCGAACUGGCAGCGGGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU ...........---........((((.(((..(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).))))))) ( -33.80) >DroEre_CAF1 47590 110 - 1 C-UGGAGGAGA---UGUGGCGAACUGGUAGCGGGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU (-((.((....---(((.((......)).)))(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).)).))). ( -33.40) >DroYak_CAF1 47359 110 - 1 C-AGAAGGAGA---UGUGGCGAACUGGCAGCGGGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU .-.........---........((((.(((..(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).))))))) ( -33.80) >DroAna_CAF1 50512 108 - 1 A-AGCAGGAG-----GAAGAACACUGGCAGCGAGCUUUUCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGUAAAGUUACUGCAGU .-.((((.((-----((....(((((((.((.((((((((((...........))))))))))))......)).)))).)....))))......((((.....))))))))... ( -30.80) >DroPer_CAF1 87466 113 - 1 G-GAAAGGCCACCGAGGAGCGAACUGCCAGCGAGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU (-(.....))............(((((.((..(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).))))))) ( -34.60) >consensus C_AGGAGGAGA___UGUGGCGAACUGGCAGCGGGCUUUGCGCUUAAGUCUAAUGCGAAAAGUUGUGAAUAGGCUCAGUAGGAAAUCCUAUUUUAUGGCGCAAAGUUACUGCAGU ......................((((.(((..(((((((((((..((((((..((((....))))...)))))).((((((....))))))....))))))))))).))))))) (-32.78 = -32.58 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:25 2006