
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,889,559 – 14,889,656 |
| Length | 97 |
| Max. P | 0.714446 |

| Location | 14,889,559 – 14,889,656 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.25 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.18 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14889559 97 - 22407834 UGGG------AGCAUGAAAUCACAGCGCAAUUGCUAUGCAAAAUAUACGUAAAUAUAAUUUACUUUAUUUUGUGGUGCUUCAUU-GCGUU-U-ACCCC---------ACCA-----UCUU ((((------....((....)).((((((((.((...((((((((...((((((...))))))..))))))))...))...)))-)))))-.-..)))---------)...-----.... ( -23.60) >DroPse_CAF1 103955 99 - 1 UGGAUGAAAAAGAAUCAAAUCACAUCGCAAUUGCUAUGCAAAGUACACGUAAAUAUAAUUUACUUUAUUUUGCGGUGCUUCAUU-GCCCUGU-ACGCC---------GCC---------- .((.((.....((......))(((..(((((.((..(((((((((...((((((...))))))..)))))))))..))...)))-))..)))-.))))---------...---------- ( -19.80) >DroSim_CAF1 46918 97 - 1 UGGG------AGCAUUAAAUCACAGCGCAAUUGCCAUGCAAAAUAUACGUAAAUAUAAUUUACUUUAUUUUGUGGUGCUUCAUU-GCGCU-U-ACCCC---------ACCA-----UCUU ((((------.(........)..((((((((.((..(((((((((...((((((...))))))..)))))))))..))...)))-)))))-.-..)))---------)...-----.... ( -23.80) >DroWil_CAF1 220240 101 - 1 UGCC------GGAAAAAAAUCACAUCGCAAUUGCUAUGCAAAAUACACGUAAAUAUAAUUUACUUUAUUUUGUGGUGCUUCAAUUGGGGG---GU-CC---------GGGAUUGGCUCCU ..((------(((.........((((((((((((...)))).......((((((...))))))......))))))))((((....)))).---.)-))---------))........... ( -22.10) >DroYak_CAF1 46554 101 - 1 UGGG--AGGGAGCUUGAAAUCACAGCGCAAUUGCUAUGCAAAAUAUACGUAAAUAUAAUUUACUUUAUUUUGUGGUGCUUCAUU-GCGUU-U-ACCCC---------GCCA-----UCUU ..((--.(((.(((((....)).)))(((((.((...((((((((...((((((...))))))..))))))))...))...)))-))...-.-..)))---------.)).-----.... ( -27.40) >DroAna_CAF1 49629 107 - 1 CGGC------GAAAUAAAAUCACAUCGCAAUUGCUAUGCAAAAUACACGUAAAUAUAAUUUACUUUAUUUUGCGGUGCUUCAUU-GCGCU-CCAGCCCGGCUCCAGUACCA-----UCCU ..((------((((((((....(((.((....)).)))..........((((((...))))))))))))))))((((((...((-(.((.-...)).)))....)))))).-----.... ( -23.80) >consensus UGGG______AGAAUAAAAUCACAGCGCAAUUGCUAUGCAAAAUACACGUAAAUAUAAUUUACUUUAUUUUGUGGUGCUUCAUU_GCGCU_U_ACCCC_________ACCA_____UCUU .......................((((((((.((..(((((((((...((((((...))))))..)))))))))..))...))).))))).............................. (-12.77 = -13.18 + 0.42)

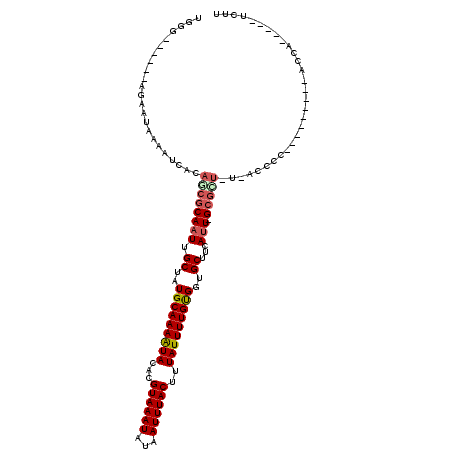
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:23 2006