| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,873,721 – 14,873,817 |
| Length | 96 |
| Max. P | 0.924443 |

| Location | 14,873,721 – 14,873,817 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -12.94 |
| Energy contribution | -13.69 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
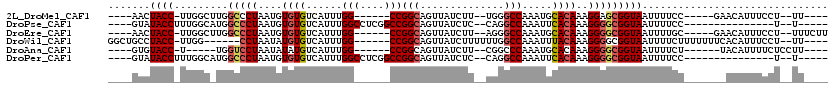
>2L_DroMel_CAF1 14873721 96 + 22407834 ----AACUACC-UUGGCUUGGCCCUAAUGUGUGUCAUUUGG------CCGGCAGUUAUCUU--UGGGCCAAAUGCACAAAGGAGCGGUAAUUUUCC-----GAACAUUUCCU--UU---- ----.......-((((....(((((....((((.(((((((------((.(.((....)).--).)))))))))))))....)).)))......))-----)).........--..---- ( -29.60) >DroPse_CAF1 82391 92 + 1 ----GUAUACCUUUGGCAUGGCCCUAAUGUGUGUCAUUUGGCCUCGGCCGGCAGUUAUCUC--CAGGCCAAAUUCACAAAGGGGCGGUAAUUUUCC---------------U--U----- ----((...((((((....((((..((((.....)))).))))..((((((.((....)))--).)))).......)))))).))((.......))---------------.--.----- ( -29.50) >DroEre_CAF1 33416 100 + 1 ----AACUACC-UUGGCUUGGCCCUAAUGUGUGUCAUUUGG------CCGGCAGUUAUCUU--AGGGCCAAAUGCACAAAGGGGCGGUAAUUUUGC-----GAACAUUUCCU--UUUCUU ----.......-..((..(((((((....((((.(((((((------((...((....)).--..)))))))))))))..))))).(((....)))-----...))...)).--...... ( -30.30) >DroWil_CAF1 196936 101 + 1 GGCUGCCUACC-UUGG------CCUAAUAUGUGUCAUUUGG------CCGGCAGUUAUCUUUUUUGGCCAAAUUUACAAAGGGGCGGUAAUUUUCUUUUUUUCACAUUUCCU--UU---- .((((((..((-((((------((......).)))((((((------((((.((......)).)))))))))).....))))))))))........................--..---- ( -25.90) >DroAna_CAF1 32411 92 + 1 ----GUGUACC-U-----UGGUCCUAAUAUAUGUCAUUUGG------CCGGCAGUUAUCUU--CGGCCCAAAUGCACAAAGGGGCGGUAAUUUUCU------UACAUUUUCUCCUU---- ----...((((-.-----..(((((......((((((((((------((((.((....)))--))).))))))).)))..))))))))).......------..............---- ( -23.40) >DroPer_CAF1 64994 92 + 1 ----GUAUACCUUUGGCAUGGCCCUAAUGUGUGUCAUUUGGCCUCGGCCGGCAGUUAUCUC--CAGGCCAAAUUCACAAAGGGGCGGUAAUUUUCC---------------U--U----- ----((...((((((....((((..((((.....)))).))))..((((((.((....)))--).)))).......)))))).))((.......))---------------.--.----- ( -29.50) >consensus ____GUCUACC_UUGGC_UGGCCCUAAUGUGUGUCAUUUGG______CCGGCAGUUAUCUU__CGGGCCAAAUGCACAAAGGGGCGGUAAUUUUCC_______ACAUUUCCU__UU____ .......((((.........(((((....((((......(((....)))(((..............))).....))))..)))))))))............................... (-12.94 = -13.69 + 0.75)
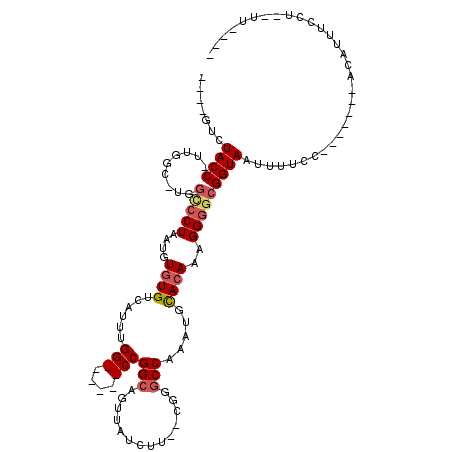
| Location | 14,873,721 – 14,873,817 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.00 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14873721 96 - 22407834 ----AA--AGGAAAUGUUC-----GGAAAAUUACCGCUCCUUUGUGCAUUUGGCCCA--AAGAUAACUGCCGG------CCAAAUGACACACAUUAGGGCCAAGCCAA-GGUAGUU---- ----..--...........-----....(((((((.......(((((((((((((..--.((....))...))------))))))).))))......(((...)))..-)))))))---- ( -28.80) >DroPse_CAF1 82391 92 - 1 -----A--A---------------GGAAAAUUACCGCCCCUUUGUGAAUUUGGCCUG--GAGAUAACUGCCGGCCGAGGCCAAAUGACACACAUUAGGGCCAUGCCAAAGGUAUAC---- -----.--.---------------((.......))((((...((((.((((((((((--(.(........)..)).)))))))))....))))...)))).(((((...)))))..---- ( -29.90) >DroEre_CAF1 33416 100 - 1 AAGAAA--AGGAAAUGUUC-----GCAAAAUUACCGCCCCUUUGUGCAUUUGGCCCU--AAGAUAACUGCCGG------CCAAAUGACACACAUUAGGGCCAAGCCAA-GGUAGUU---- ......--...........-----....(((((((((((((.(((((((((((((..--.((....))...))------))))))).))))....))))....))...-)))))))---- ( -29.90) >DroWil_CAF1 196936 101 - 1 ----AA--AGGAAAUGUGAAAAAAAGAAAAUUACCGCCCCUUUGUAAAUUUGGCCAAAAAAGAUAACUGCCGG------CCAAAUGACACAUAUUAGG------CCAA-GGUAGGCAGCC ----..--.((...(((.............((((((((....(((..((((((((.....((....))...))------))))))...))).....))------)...-)))))))).)) ( -20.11) >DroAna_CAF1 32411 92 - 1 ----AAGGAGAAAAUGUA------AGAAAAUUACCGCCCCUUUGUGCAUUUGGGCCG--AAGAUAACUGCCGG------CCAAAUGACAUAUAUUAGGACCA-----A-GGUACAC---- ----..............------.......((((...(((.(((((((((((.(((--.((....))..)))------))))))).))))....)))....-----.-))))...---- ( -22.90) >DroPer_CAF1 64994 92 - 1 -----A--A---------------GGAAAAUUACCGCCCCUUUGUGAAUUUGGCCUG--GAGAUAACUGCCGGCCGAGGCCAAAUGACACACAUUAGGGCCAUGCCAAAGGUAUAC---- -----.--.---------------((.......))((((...((((.((((((((((--(.(........)..)).)))))))))....))))...)))).(((((...)))))..---- ( -29.90) >consensus ____AA__AGGAAAUGU_______GGAAAAUUACCGCCCCUUUGUGAAUUUGGCCCG__AAGAUAACUGCCGG______CCAAAUGACACACAUUAGGGCCA_GCCAA_GGUAGAC____ ...............................((((((((...((((.....(((..............)))(((....)))........))))...)))).........))))....... (-11.91 = -12.44 + 0.53)
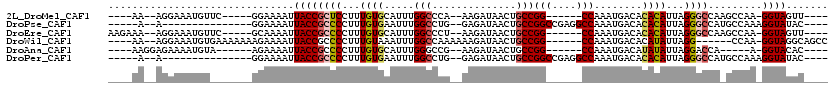
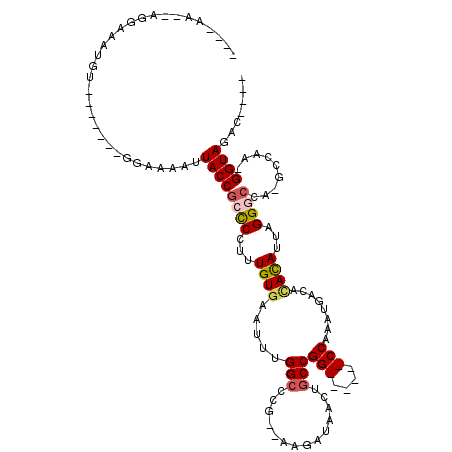
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:19 2006