
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,866,190 – 14,866,281 |
| Length | 91 |
| Max. P | 0.999845 |

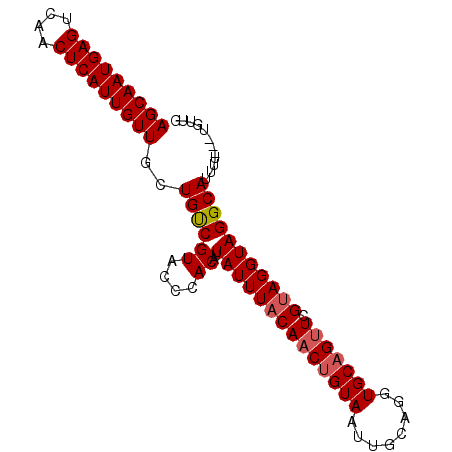
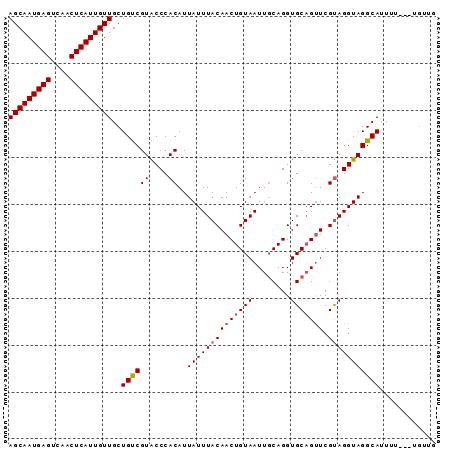
| Location | 14,866,190 – 14,866,281 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -22.74 |
| Energy contribution | -23.18 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14866190 91 + 22407834 AGCAAUGAGUCAACUCAUUGUUGCUGUCGUACCCACAUUAUUUACAACUGUAAUUGCAGGUGCAGUUCGUAGGUAGGCAUUUUUUUUUUUG (((((((((....)))))))))..((((((....))..((((((((((((((........))))))).)))))))))))............ ( -24.40) >DroSec_CAF1 26047 88 + 1 AGCAAUGAGUCAACUCAUUGUUGCUGUCGUACCCACAUUAUUUACAACUGUAAUUGCAGGUGCAGCUCGUAGGUAGGCAUUUU---UGUUG (((((((((....)))))))))..((((.((((.((.....((((....))))((((....))))...)).))))))))....---..... ( -23.70) >DroSim_CAF1 25566 88 + 1 AGCAAUGAGUCAACUCAUUGUUGCUGUCGUACCCACAUUAUUUACAACUGUAAUUUCAGGUGCUGUUCGUAGGUAGGCAUUUU---UGUUG (((((((((....)))))))))..((((.((((.((...((...((.(((......))).))..))..)).))))))))....---..... ( -20.90) >DroEre_CAF1 25444 86 + 1 AGCAAUGAGUCAACUCAUUGUUGCUGCCGUGCCCACAUUAUUUACAACUGUAAUUGCAGGUGCAGUUCGCAGGUAGGCAGU-----GGUUG (((((((((....)))))))))((((((((....))...........((((((((((....)))))).))))...))))))-----..... ( -31.50) >DroYak_CAF1 25199 86 + 1 AGCAAUGAGUCAACUCAUUGUUGCUGUCGUGCCCACAUUAUUUACAACUGUAAUUGCAGGUGCAGUUCGUAGGUAGGCAUG-----GGUUG (((((((((....)))))))))....(((((((.....((((((((((((((........))))))).)))))))))))))-----).... ( -30.50) >consensus AGCAAUGAGUCAACUCAUUGUUGCUGUCGUACCCACAUUAUUUACAACUGUAAUUGCAGGUGCAGUUCGUAGGUAGGCAUUUU___UGUUG (((((((((....)))))))))..((((((....))..((((((((((((((........))))))).)))))))))))............ (-22.74 = -23.18 + 0.44)

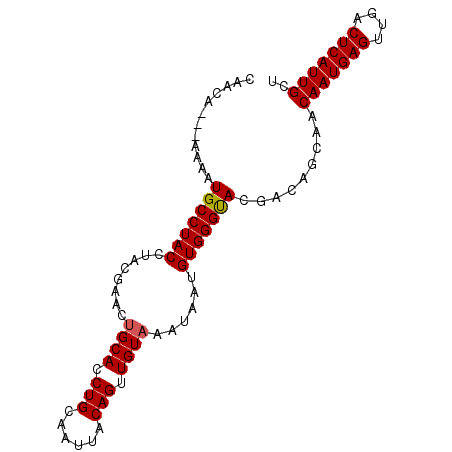
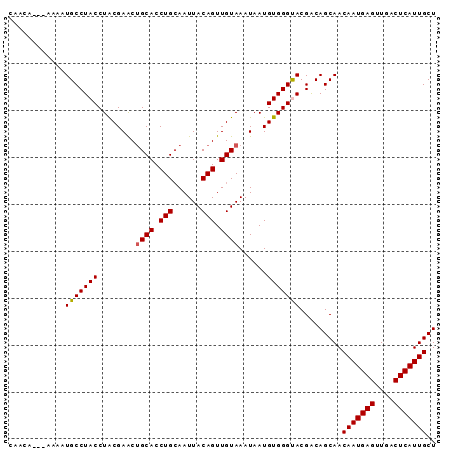
| Location | 14,866,190 – 14,866,281 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -22.93 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14866190 91 - 22407834 CAAAAAAAAAAAUGCCUACCUACGAACUGCACCUGCAAUUACAGUUGUAAAUAAUGUGGGUACGACAGCAACAAUGAGUUGACUCAUUGCU ............(((.((((((((...((((.(((......))).)))).....)))))))).....))).(((((((....))))))).. ( -22.50) >DroSec_CAF1 26047 88 - 1 CAACA---AAAAUGCCUACCUACGAGCUGCACCUGCAAUUACAGUUGUAAAUAAUGUGGGUACGACAGCAACAAUGAGUUGACUCAUUGCU .....---....(((.((((((((...((((.(((......))).)))).....)))))))).....))).(((((((....))))))).. ( -22.50) >DroSim_CAF1 25566 88 - 1 CAACA---AAAAUGCCUACCUACGAACAGCACCUGAAAUUACAGUUGUAAAUAAUGUGGGUACGACAGCAACAAUGAGUUGACUCAUUGCU .....---....(((.((((((((....(((.(((......))).)))......)))))))).....))).(((((((....))))))).. ( -21.30) >DroEre_CAF1 25444 86 - 1 CAACC-----ACUGCCUACCUGCGAACUGCACCUGCAAUUACAGUUGUAAAUAAUGUGGGCACGGCAGCAACAAUGAGUUGACUCAUUGCU .....-----.(((((..((..((...((((.(((......))).)))).....))..))...)))))...(((((((....))))))).. ( -26.20) >DroYak_CAF1 25199 86 - 1 CAACC-----CAUGCCUACCUACGAACUGCACCUGCAAUUACAGUUGUAAAUAAUGUGGGCACGACAGCAACAAUGAGUUGACUCAUUGCU .....-----..(((((((........((((.(((......))).))))......))))))).........(((((((....))))))).. ( -22.14) >consensus CAACA___AAAAUGCCUACCUACGAACUGCACCUGCAAUUACAGUUGUAAAUAAUGUGGGUACGACAGCAACAAUGAGUUGACUCAUUGCU ............(((((((........((((.(((......))).))))......))))))).........(((((((....))))))).. (-20.70 = -20.66 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:15 2006