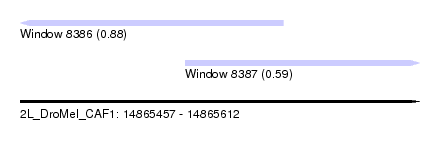
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,865,457 – 14,865,612 |
| Length | 155 |
| Max. P | 0.875169 |

| Location | 14,865,457 – 14,865,559 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 75.57 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -19.67 |
| Energy contribution | -20.45 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
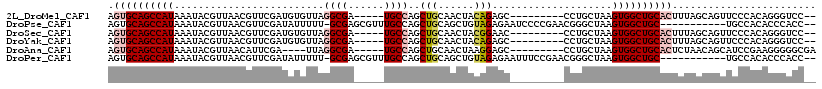
>2L_DroMel_CAF1 14865457 102 - 22407834 AGUGCAGCCAUAAAUACGUUAACGUUCGAUGUGUUAGGCGA-----UGCCAGCUGCAACUACAGAGC---------CCUGCUAAGUGGCUGCACUUUAGCAGUUCCCACAGGGUCC-- ((((((((....((((((((.......)))))))).(((..-----.))).))))).))).....((---------((((....(..(((((......)))))..)..))))))..-- ( -34.90) >DroPse_CAF1 60876 104 - 1 AGUGCAGCCAUAAAUACGUUAACGUUCGAUAUUUUU-GCGAGCGUUUGCCAGCUGCAGCUGUAGAGAAUCCCCGAACGGGCUAAGUGGCUGC-----------UGCCACACCCACC-- ...((((((((......((.((((((((.((....)-))))))))).))((((....)))).........(((....)))....))))))))-----------.............-- ( -33.30) >DroSec_CAF1 25322 102 - 1 AGUGCAGCCAUAAAUACGUUAACGUUCGAUGUGUUAGGCGA-----UGCCAGCUGCAACUACGGAAC---------CCUGCUAAGUGGCUGCACUUUAGCAGUUCCCACAGGGUCC-- ((((((((....((((((((.......)))))))).(((..-----.))).))))).)))..(((.(---------((((....(..(((((......)))))..)..))))))))-- ( -36.90) >DroYak_CAF1 24448 102 - 1 AGUGCAGCCAUAAAUACGUUAACGUUCGAUGUGUUAGGCGA-----UGCCAGCUGCAACUACAGAGC---------CCUGCUAAGUGGCUGCACUUUAGCAGUUCCCACAGGGUCC-- ((((((((....((((((((.......)))))))).(((..-----.))).))))).))).....((---------((((....(..(((((......)))))..)..))))))..-- ( -34.90) >DroAna_CAF1 22884 100 - 1 AGUGCAGCCAUAAAUACGUUAACAUUCGA----UUAGGCGA-----UGCCAGCUGCAACUAAGGAGC---------CCUGCUAAGUGGCUGCACUCUAACAGCAUCCGAAGGGGGCGA (((((((((((..................----...(((..-----.))).(((.(......).)))---------........)))))))))))......((.(((....))))).. ( -33.10) >DroPer_CAF1 49286 104 - 1 AGUGCAGCCAUAAAUACGUUAACGUUCGAUAUUUUU-GCGAGCGUUUGCCAGCUGCAGCUGUAGAGAAUUUCCGAACGGGCUAAGUGGCUGC-----------UGCCACACCCACC-- .(((.((((........((.((((((((.((....)-))))))))).))((((....)))).................))))..(((((...-----------.)))))...))).-- ( -32.20) >consensus AGUGCAGCCAUAAAUACGUUAACGUUCGAUGUGUUAGGCGA_____UGCCAGCUGCAACUACAGAGC_________CCUGCUAAGUGGCUGCACUUUAGCAGUUCCCACAGGGUCC__ .((((((((((.........................((((......))))..(((......)))....................))))))))))........................ (-19.67 = -20.45 + 0.78)

| Location | 14,865,521 – 14,865,612 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 92.58 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -23.01 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
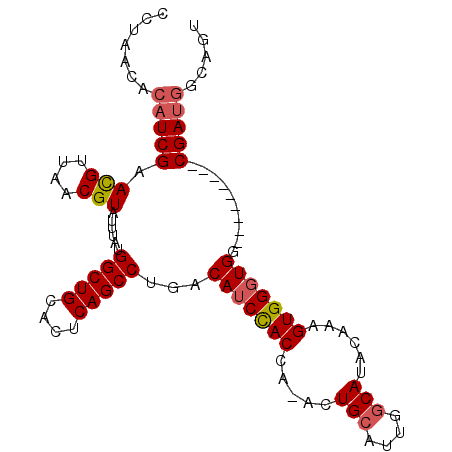
>2L_DroMel_CAF1 14865521 91 + 22407834 CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCA-ACUGCAUUGGCAUACAAAGUGGGUGG---------CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((..-..(((....)))......))))))).---------)))))..... ( -26.00) >DroSec_CAF1 25386 91 + 1 CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCA-ACUGCAUUGGCAUACAAAGUGGGUGG---------CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((..-..(((....)))......))))))).---------)))))..... ( -26.00) >DroSim_CAF1 24905 91 + 1 CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCA-ACUGCAUUGGCAUACAAAGUGGGUGG---------CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((..-..(((....)))......))))))).---------)))))..... ( -26.00) >DroEre_CAF1 24747 92 + 1 CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCAAACUGCAUUGGCAUACAAAGUGGGUGG---------CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((.....(((....)))......))))))).---------)))))..... ( -26.40) >DroYak_CAF1 24512 91 + 1 CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCA-ACUGCAUUGGCAUACAAAGUGGGUGG---------CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((..-..(((....)))......))))))).---------)))))..... ( -26.00) >DroAna_CAF1 22950 96 + 1 CCUAA----UCGAAUGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAGCUACCA-ACUGCAAUGCCACAAAAAGUGGGUGGGCUGUGUGACGAUGGCAGU .....----.....(((((.(((......(((((....)))))..(((((((((.-(((..............))).))).))))))...))).))))).. ( -26.14) >consensus CCUAACACAUCGAACGUUAACGUAUUUAUGGCUGCACUCAGCCUGACAUCCACCA_ACUGCAUUGGCAUACAAAGUGGGUGG_________CGAUGGCAGU .......(((((.(((....)))......(((((....)))))...(((((((.....(((....)))......)))))))..........)))))..... (-23.01 = -23.40 + 0.39)

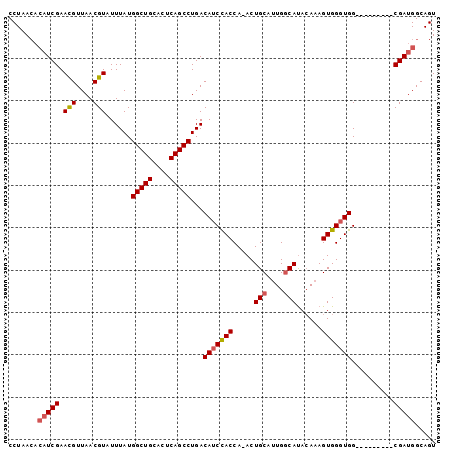
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:13 2006