| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,609,472 – 1,609,587 |
| Length | 115 |
| Max. P | 0.911716 |

| Location | 1,609,472 – 1,609,587 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -45.34 |
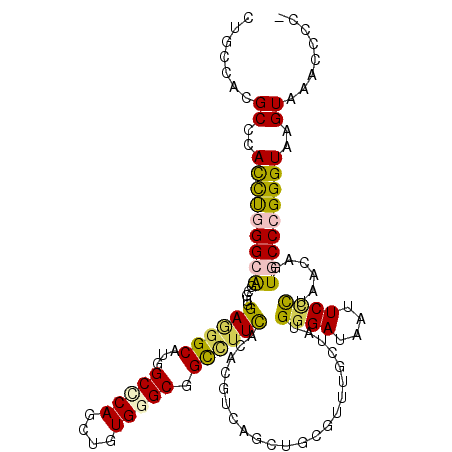
| Consensus MFE | -27.09 |
| Energy contribution | -27.09 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
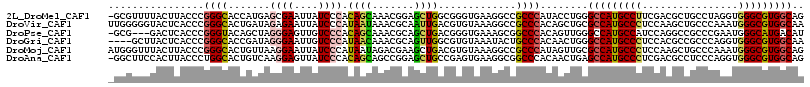
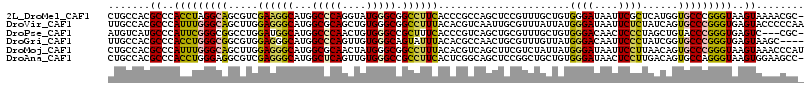
>2L_DroMel_CAF1 1609472 115 + 22407834 -GCGUUUUACUUACCCGGGCACCAUGAGCGAAUUAUCCCACAGCAAACGGAGCUGGCGGGUGAAGGCCGCCCAUACCUGGGCCAUGCCUUCGACGCUGCCUAGGUGGGCGUGGCAG -..(((...((((((..((((......(((......(((.((((.......))))..)))(((((((.(((((....)))))...))))))).)))))))..))))))...))).. ( -48.30) >DroVir_CAF1 130331 116 + 1 UUGGGGGUACUCACCCGGGCACUGAUAGAGAAUUAUCCCAUAAUAAACGCAAUUGACGUGUAAAGGCCGCCCACAGCUGCGCCAUGCCCUCCAAGCUGCCCAAAUGGGCGUGGCAA ((((((((....))))(((((..(((((....))))).........((((.......))))...(((.((........))))).))))).))))(((((((....))))..))).. ( -36.20) >DroPse_CAF1 136359 112 + 1 -GCG---GACUCACCCGGGUACAGCUAGGGAGUUGUCCCACAGCAAACGCAGCUGACGGGUGAAAGCGGCCCACAGUUGGGCCAUGCCAUCCAGGCCGCCCGAAUGGGCAUGACAU -((.---...(((((((((.((((((....))))))))).((((.......))))..))))))..)).(((((...((((((...(((.....))).)))))).)))))....... ( -53.10) >DroGri_CAF1 114750 112 + 1 ----GCUUACUCACCCGGGCACCGAUAGGGAAUUGUCCCAUAACAAACGCAGUUGGCGUGUAAAUACUGCCCACAACUGGGCCAUGCCCUCCACGCCGCCCAGGUGGGCGUGGCAA ----(((..((((((.((((.......((((....))))...............((((((........(((((....))))).........)))))))))).))))))...))).. ( -46.93) >DroMoj_CAF1 135263 116 + 1 AUGGGUUUACUUACCCGGGCACUGUUAAGGAAUUAUCCCAUAAUAGACGAAGCUGACGUGUAAAGGCCGCCCAUAGUUGCGCCAUGCCCUCCAAGCUGCCCAAAUGGGCGUGGCAG ..((((......))))((((.((((((.((.......)).)))))).....(((..........))).))))........(((((((((................))))))))).. ( -40.79) >DroAna_CAF1 117987 115 + 1 -GGCUUCCACUUACCCUGGCACUGUCAAGGAGUUAUCCCACAGCAGCCGGAGCUGCCGAGUGAAGGCGGCCCACAACUGAGCCAUGCCCUCGACGCCUCCCAGGUGGGCGUGGCAG -((((.(((.......)))..((((...(((....))).)))).))))((.((((((.......))))))))........(((((((((.....(((.....)))))))))))).. ( -46.70) >consensus _GGGGUUUACUCACCCGGGCACUGUUAGGGAAUUAUCCCACAACAAACGCAGCUGACGUGUAAAGGCCGCCCACAGCUGGGCCAUGCCCUCCACGCCGCCCAAAUGGGCGUGGCAG ................((((.......((((....)))).((((.......)))).............))))........(((((((((................))))))))).. (-27.09 = -27.09 + 0.00)
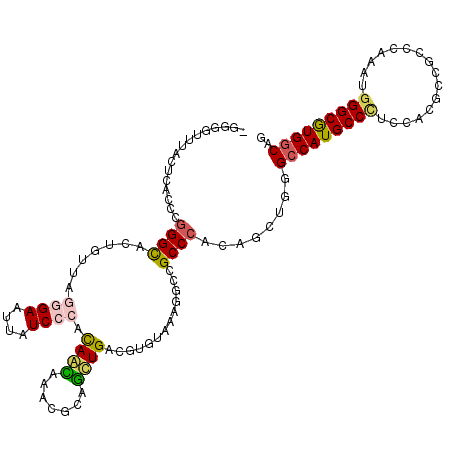
| Location | 1,609,472 – 1,609,587 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.78 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -28.43 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1609472 115 - 22407834 CUGCCACGCCCACCUAGGCAGCGUCGAAGGCAUGGCCCAGGUAUGGGCGGCCUUCACCCGCCAGCUCCGUUUGCUGUGGGAUAAUUCGCUCAUGGUGCCCGGGUAAGUAAAACGC- .((((.((..((((...(.((((..((((((...(((((....))))).)))))).(((((.(((.......))))))))......)))))..))))..))))))..........- ( -49.90) >DroVir_CAF1 130331 116 - 1 UUGCCACGCCCAUUUGGGCAGCUUGGAGGGCAUGGCGCAGCUGUGGGCGGCCUUUACACGUCAAUUGCGUUUAUUAUGGGAUAAUUCUCUAUCAGUGCCCGGGUGAGUACCCCCAA .......((((....))))....(((.(((((((((((((.....((((.........))))..)))))))......((((....)))).....))))))((((....))))))). ( -39.80) >DroPse_CAF1 136359 112 - 1 AUGUCAUGCCCAUUCGGGCGGCCUGGAUGGCAUGGCCCAACUGUGGGCCGCUUUCACCCGUCAGCUGCGUUUGCUGUGGGACAACUCCCUAGCUGUACCCGGGUGAGUC---CGC- .....(((((.(((((((...))))))))))))((((((....))))))(..((((((((.(((((((....))...((((....)))).)))))....))))))))..---)..- ( -50.80) >DroGri_CAF1 114750 112 - 1 UUGCCACGCCCACCUGGGCGGCGUGGAGGGCAUGGCCCAGUUGUGGGCAGUAUUUACACGCCAACUGCGUUUGUUAUGGGACAAUUCCCUAUCGGUGCCCGGGUGAGUAAGC---- .......((.(((((((((((((((....((...(((((....))))).)).....))))))...............((((....)))).......))))))))).))....---- ( -47.30) >DroMoj_CAF1 135263 116 - 1 CUGCCACGCCCAUUUGGGCAGCUUGGAGGGCAUGGCGCAACUAUGGGCGGCCUUUACACGUCAGCUUCGUCUAUUAUGGGAUAAUUCCUUAACAGUGCCCGGGUAAGUAAACCCAU ..((((.((((.((..((...))..)))))).))))........(((((((........)))........((.(((.((((....))))))).)).))))((((......)))).. ( -37.70) >DroAna_CAF1 117987 115 - 1 CUGCCACGCCCACCUGGGAGGCGUCGAGGGCAUGGCUCAGUUGUGGGCCGCCUUCACUCGGCAGCUCCGGCUGCUGUGGGAUAACUCCUUGACAGUGCCAGGGUAAGUGGAAGCC- ((.((((((((.....((((..(((((((((..((((((....)))))))))....))))))..))))(((.(((((((((....))))..)))))))).))))..)))).))..- ( -52.30) >consensus CUGCCACGCCCACCUGGGCAGCGUGGAGGGCAUGGCCCAGCUGUGGGCGGCCUUCACACGUCAGCUGCGUUUGCUAUGGGAUAAUUCCCUAACAGUGCCCGGGUAAGUAAACCCC_ .......((..(((((((((.....((((((...(((((....))))).))))))......................((((....))))......)))))))))..))........ (-28.43 = -28.02 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:23 2006