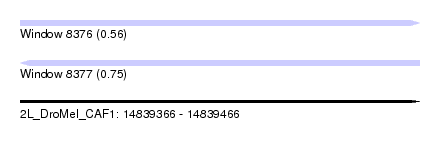
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,839,366 – 14,839,466 |
| Length | 100 |
| Max. P | 0.745677 |

| Location | 14,839,366 – 14,839,466 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -17.02 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.83 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
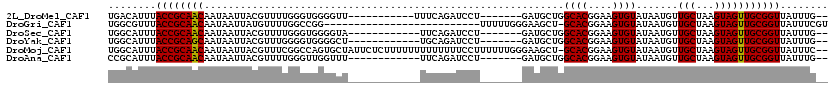
>2L_DroMel_CAF1 14839366 100 + 22407834 --CAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGCCAGCAUC-------AGGAUCUGAAA-----------AACCCCACCCAAAACGUAAUUAUUGUUGCGGUAAAUGUCA --......((((((((.................((..((((((....))).-------.)))..))...-----------......((.......)).......))))))))........ ( -16.30) >DroGri_CAF1 100097 93 + 1 ACGAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGC-AGCUUCCCAAAAA--------------------------CCGGCCAAAACAUAAUUAUUGUUGCGGUAAACGCCA ........((((((((.................(((....))).-.(((.........--------------------------..)))...............))))))))........ ( -14.60) >DroSec_CAF1 81203 99 + 1 --CAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGCCAGCAUC-------AGGAUCUGAA------------UACCCCACCCAAAACGUAAUUAUUGUUGCGGUAAAUGCCA --......((((((((.................((..((((((....))).-------.)))..))..------------(((............)))......))))))))........ ( -16.90) >DroYak_CAF1 78752 99 + 1 --CAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGCCAGCAUC-------AGGAUCUGCA------------AGCCCCACCCCAAACGUAAUUAUUGCUGCGGUAAAUGCCA --.........(((..((((((((((.(((((.(((....)))...(((..-------......))).------------...............)))))..)))))).))))..))).. ( -15.50) >DroMoj_CAF1 119572 117 + 1 --GAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGC-AGCUUCCCAAAAAAGGAAAAAAAAAAAAAGAGAAUAGCACUGGCCGAAACGUAAUUAUUGUUGCGGUAAAUGCCA --......((((((((......((.........(((....))).-...((((.......))))..................))....((......)).......))))))))........ ( -17.90) >DroAna_CAF1 75563 99 + 1 --CAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGCCAGCAUC-------AGGAUCUGAA------------AAACCAACCCAAAACGUAAUUAUUGUUGCGGUAAAUGCGG --.......(((((..((((..((((((.(((((...((((((....))).-------.)))..((..------------....)).........)))..)).))))))))))..))))) ( -20.90) >consensus __CAAAUAACCGCAACUACUUAGCAACAUUAUACACUUCCGUGCCAGCAUC_______AGGAUCUGAA____________AACCCCACCCAAAACGUAAUUAUUGUUGCGGUAAAUGCCA ........((((((((.................(((....)))................((...........................))..............))))))))........ (-11.33 = -11.83 + 0.50)



| Location | 14,839,366 – 14,839,466 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.95 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -18.91 |
| Energy contribution | -18.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
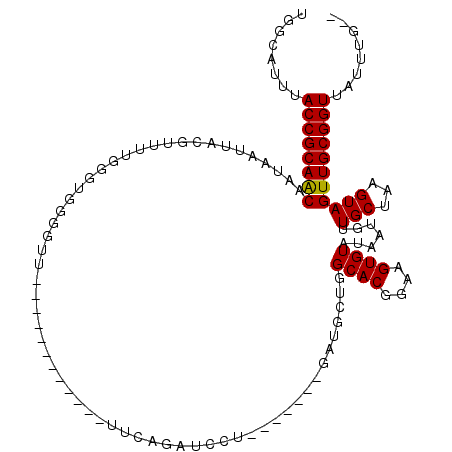
>2L_DroMel_CAF1 14839366 100 - 22407834 UGACAUUUACCGCAACAAUAAUUACGUUUUGGGUGGGGUU-----------UUUCAGAUCCU-------GAUGCUGGCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUG-- ...((...((((((((........((((..((((..((..-----------..))..)))).-------))))...((((....))))................))))))))....))-- ( -26.50) >DroGri_CAF1 100097 93 - 1 UGGCGUUUACCGCAACAAUAAUUAUGUUUUGGCCGG--------------------------UUUUUGGGAAGCU-GCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUCGU ..(((...((((((((((((....))))(((((.((--------------------------((((...))))))-((((....))))........)))))...)))))))).....))) ( -26.50) >DroSec_CAF1 81203 99 - 1 UGGCAUUUACCGCAACAAUAAUUACGUUUUGGGUGGGGUA------------UUCAGAUCCU-------GAUGCUGGCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUG-- ........((((((((......(((......((..(((((------------(.(((...))-------)))))).((((....))))......)..))..)))))))))))......-- ( -25.40) >DroYak_CAF1 78752 99 - 1 UGGCAUUUACCGCAGCAAUAAUUACGUUUGGGGUGGGGCU------------UGCAGAUCCU-------GAUGCUGGCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUG-- ........((((((((...........(..((((..(...------------..)..)))).-------.)((((.((((....))))............))))))))))))......-- ( -26.62) >DroMoj_CAF1 119572 117 - 1 UGGCAUUUACCGCAACAAUAAUUACGUUUCGGCCAGUGCUAUUCUCUUUUUUUUUUUUUCCUUUUUUGGGAAGCU-GCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUC-- ..(((.((((.((((((........((((((((....)))...................((......)))))))(-((((....)))))...))))))...)))).))).........-- ( -28.70) >DroAna_CAF1 75563 99 - 1 CCGCAUUUACCGCAACAAUAAUUACGUUUUGGGUUGGUUU------------UUCAGAUCCU-------GAUGCUGGCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUG-- (((((.((((.((((((.....((((((((.(((..((..------------.((((...))-------)).))..)).).))))))))...))))))...)))).))))).......-- ( -31.80) >consensus UGGCAUUUACCGCAACAAUAAUUACGUUUUGGGUGGGGUU____________UUCAGAUCCU_______GAUGCUGGCACGGAAGUGUAUAAUGUUGCUAAGUAGUUGCGGUUAUUUG__ ........((((((((............................................................((((....)))).......(((...)))))))))))........ (-18.91 = -18.77 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:02 2006