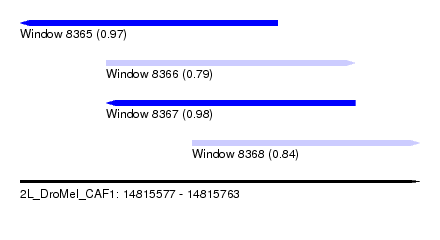
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,815,577 – 14,815,763 |
| Length | 186 |
| Max. P | 0.979237 |

| Location | 14,815,577 – 14,815,697 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -26.57 |
| Energy contribution | -27.43 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14815577 120 - 22407834 UUUACUUUCGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGGUCCCCAAAUCAAAGGGGAGU (((((((((.(((((.....(..((((.....))))..)......))))).)))))))))............(((.((((((........)))))).)))(((((........))))).. ( -33.20) >DroSec_CAF1 57060 119 - 1 UUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGGUCCCCA-AUCAAAGGGGAGU (((((((((.(((((.....(..((((.....))))..)......))))).)))))))))............(((.((((((........)))))).)))(((((.-......))))).. ( -31.40) >DroSim_CAF1 57621 120 - 1 UUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGGUCCCCAAAUCAAAGGGGAGU (((((((((.(((((.....(..((((.....))))..)......))))).)))))))))............(((.((((((........)))))).)))(((((........))))).. ( -31.70) >DroEre_CAF1 52877 120 - 1 UUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGACUGGCGGAAAGUAAAGAAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGGUCCCCAAAUCAAAGGGGAGU (((((((((.((((......(..((((.....))))..).......)))).)))))))))............(((.((((((........)))))).)))(((((........))))).. ( -28.62) >DroYak_CAF1 54488 120 - 1 UUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGAAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGUUCCCCAAAUCAAAGGGGAGU (((((((((.(((((.....(..((((.....))))..)......))))).)))))))))(((((....))))).........................((((((........)))))). ( -32.00) >DroAna_CAF1 52704 118 - 1 UUUACUUUUGGUCACAGAAACUUCUGCAGAUGGCAGCUGGCAGGGCUGGGGGAAAGUAAAGCAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUU--UGCUACCGAAGUCAAAGGGGAGC ....((((((.........(((((.((((((..(((((.....)))))(..(((.((........)).)))..)..................)))--)))....)))))))))))..... ( -21.80) >consensus UUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAUUUAAAAUUGCAAAUUUUUGGUCCCCAAAUCAAAGGGGAGU (((((((((.(((((.....(..((((.....))))..)......))))).))))))))).............((.((((((........)))))).)).(((((........))))).. (-26.57 = -27.43 + 0.86)


| Location | 14,815,617 – 14,815,733 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14815617 116 + 22407834 AUUUAUGAAAAAUGUUUUGCUUUACUUUCCGCCACUCUUGCCAGCUGCCAACUGCAGAAGUUUUUGUGACCGAAAGUAAAGUUUGUCAAAACAAAAAAAA----AAAAAGAGAAGAAACC ............(((((((((((((((((.(.(((....((...((((.....))))..))....))).).)))))))))).....))))))).......----................ ( -25.40) >DroSim_CAF1 57661 120 + 1 AUUUAUGAAAAAUGUUUUGCUUUACUUUCCGCCACUCUUGCCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAAACAAAAAAGAAAAGAAACC ............((((((((((((((((..(.(((....((...((((.....))))..))....))).)..))))))))))((((....))))....))))))................ ( -22.00) >DroEre_CAF1 52917 106 + 1 AUUUAUGAAAAAUGUUUUUCUUUACUUUCCGCCAGUCUUGCCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAACAAAA--------------AAUCC ............((((((.(((((((((..(((((....((...((((.....))))..))..))).).)..)))))))))......))))))........--------------..... ( -15.30) >DroYak_CAF1 54528 106 + 1 AUUUAUGAAAAAUGUUUUUCUUUACUUUCCGCCACUCUUGCCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAA--------------UGUCC ...........(((((((((((((((((..(.(((....((...((((.....))))..))....))).)..)))))))))(((((....))))).)))))--------------))).. ( -19.10) >DroAna_CAF1 52742 103 + 1 AUUUAUGAAAAAUGUUUUGCUUUACUUUCCCCCAGCCCUGCCAGCUGCCAUCUGCAGAAGUUUCUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAA-----------------AUC ............((((((((((((((((....((((.......))))......(((((....))))).....))))))))).....))))))).......-----------------... ( -19.90) >consensus AUUUAUGAAAAAUGUUUUGCUUUACUUUCCGCCACUCUUGCCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAA______________AAACC ............((((((((((((((((..(.(((....((...((((.....))))..))....))).)..))))))))).....)))))))........................... (-17.68 = -18.68 + 1.00)

| Location | 14,815,617 – 14,815,733 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -24.54 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
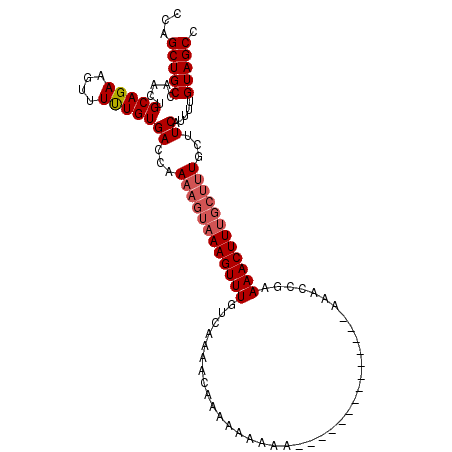
>2L_DroMel_CAF1 14815617 116 - 22407834 GGUUUCUUCUCUUUUU----UUUUUUUUGUUUUGACAAACUUUACUUUCGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAU ................----.......(((((((.....((((((((((.(((((.....(..((((.....))))..)......))))).)))))))))))))))))............ ( -29.90) >DroSim_CAF1 57661 120 - 1 GGUUUCUUUUCUUUUUUGUUUUUUUUUUGUUUUGACAAACUUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAU ...........................(((((((.....((((((((((.(((((.....(..((((.....))))..)......))))).)))))))))))))))))............ ( -28.40) >DroEre_CAF1 52917 106 - 1 GGAUU--------------UUUUGUUUUGUUUUGACAAACUUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGACUGGCGGAAAGUAAAGAAAAACAUUUUUCAUAAAU .((..--------------...((((((((....))...((((((((((.((((......(..((((.....))))..).......)))).)))))))))).))))))....))...... ( -24.42) >DroYak_CAF1 54528 106 - 1 GGACA--------------UUUUUUUUUGUUUUGACAAACUUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGAAAAACAUUUUUCAUAAAU .....--------------........((((((......((((((((((.(((((.....(..((((.....))))..)......))))).)))))))))).))))))............ ( -26.00) >DroAna_CAF1 52742 103 - 1 GAU-----------------UUUUUUUUGUUUUGACAAACUUUACUUUUGGUCACAGAAACUUCUGCAGAUGGCAGCUGGCAGGGCUGGGGGAAAGUAAAGCAAAACAUUUUUCAUAAAU ...-----------------.......(((((((....(((((.((((..(((.(((......((((.....)))))))....)))..)))))))))....)))))))............ ( -24.90) >consensus GGUUU______________UUUUUUUUUGUUUUGACAAACUUUACUUUUGGUCACAAAAACUUCUGCAGUUGGCAGCUGGCAAGAGUGGCGGAAAGUAAAGCAAAACAUUUUUCAUAAAU ...........................(((((((.....((((((((((.(((((.....(..((((.....))))..)......))))).)))))))))))))))))............ (-24.54 = -25.38 + 0.84)



| Location | 14,815,657 – 14,815,763 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.78 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -16.16 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14815657 106 + 22407834 CCAGCUGCCAACUGCAGAAGUUUUUGUGACCGAAAGUAAAGUUUGUCAAAACAAAAAAAA----AAAAAGAGAAGAAACCGAAAACUUUGCUUUGCUUCAUUUUGUAGCC ...(((((.((((.....))))...((((...((((((((((((................----..................))))))))))))...))))...))))). ( -18.28) >DroSim_CAF1 57701 110 + 1 CCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAAACAAAAAAGAAAAGAAACCGAAAACUUUGCUUUGCUUCAUUUUGUAGCC ...(((((.((((.....))))...((((...((((((((((((......................................))))))))))))...))))...))))). ( -19.26) >DroEre_CAF1 52957 96 + 1 CCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAACAAAA--------------AAUCCGAAAACUUUGCUUUGCUUCAUUUUGUAGCC ...(((((.((((.....))))...((((...((((((((((((.................--------------.......))))))))))))...))))...))))). ( -19.76) >DroYak_CAF1 54568 96 + 1 CCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAA--------------UGUCCCAAAACUUUGCUUUGCUUCAUUUUGUAGCC ...(((((.((((.....))))...((((...((((((((((((......(((........--------------)))....))))))))))))...))))...))))). ( -21.00) >DroAna_CAF1 52782 88 + 1 CCAGCUGCCAUCUGCAGAAGUUUCUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAA-----------------AUCGAAAACUU-----UGCUUCAUUUUGUAGCC ...(((((.....(((((....)))))......(((((((((((................-----------------.....))))))-----)))))......))))). ( -18.80) >consensus CCAGCUGCCAACUGCAGAAGUUUUUGUGACCAAAAGUAAAGUUUGUCAAAACAAAAAAAAA______________AAACCGAAAACUUUGCUUUGCUUCAUUUUGUAGCC ...(((((.....(((((....)))))((...((((((((((((......................................))))))))))))...)).....))))). (-16.16 = -17.00 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:54 2006