
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
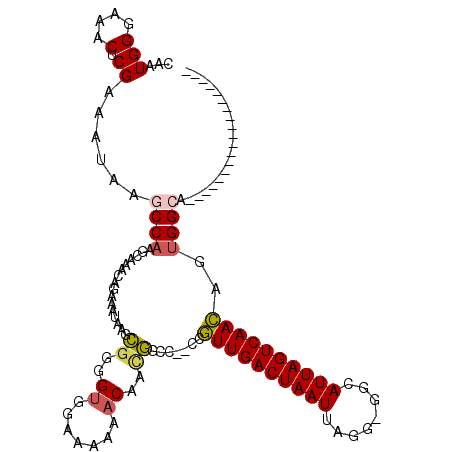
| Location | 14,814,729 – 14,814,827 |
| Length | 98 |
| Max. P | 0.772447 |

| Location | 14,814,729 – 14,814,827 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.35 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -16.83 |
| Energy contribution | -17.00 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14814729 98 + 22407834 CAAUGGGAAACUCGAAAUAAGCCAAGCAAACAGAAAUAAGCGAGGUGGAAAAAACAACGGCC--CCGUUGACUAAUUAGG-GGCAUUAGUCAACAGUGGCA------------------- ....((....))........((((.................(.(((.(.........).)))--.)((((((((((....-...))))))))))..)))).------------------- ( -22.90) >DroSec_CAF1 56218 98 + 1 CAAUGGGAAACUCGAAAUAAUCCAAGCAAACAGAAAUAACCGGGGUGGAAAAAACAACGGCC--CCGUUGACUAAUUAGG-GGCAUUAGUCAACAGUGGCA------------------- ....((....)).............((..............(((((.(.........).)))--))((((((((((....-...))))))))))....)).------------------- ( -25.60) >DroSim_CAF1 56778 98 + 1 CAAUGGGAAACUCGAAAUAAGCCAAGCAAACAGAAAUAAGCGGGGUGGAAAAAACAACGGCC--CCGUUGACUAAUUAGG-GGCAUUAGUCAACAGUGGCA------------------- ....((....))........((((.................(((((.(.........).)))--))((((((((((....-...))))))))))..)))).------------------- ( -29.80) >DroEre_CAF1 52014 98 + 1 CAAUGGGAAACUCGAAAUAAGCCAAGCAAACAGAAAUUAGCGGGGUGGAAAAAACAAUGGCC--CCGUUGACUAAUUAGG-GGCAUUAGUCAACAGUGGCA------------------- ....((....))........((((.................(((((.(.........).)))--))((((((((((....-...))))))))))..)))).------------------- ( -27.90) >DroYak_CAF1 53628 98 + 1 CAAUGGGAAACUCGAAAUAAGCCAAGCAAACAGAAAUAAGCGGGGUGGAAAAAACAACGGCC--CCGUUGACUAAUUAGG-GGCAUUAGUCAACAGUGGCA------------------- ....((....))........((((.................(((((.(.........).)))--))((((((((((....-...))))))))))..)))).------------------- ( -29.80) >DroAna_CAF1 51749 117 + 1 CAAUGGGAAGCACGAAAUGAACCCAGCAGAAA-AAACAAUGAGGGUGAAAAA--CAGCCACUGGGCUUUGACUAAUUAGAAGGCAUUAGUCAAAACAGGCUCUGUCCGGUCCCGGCCCGG ...((((...((.....))..)))).......-.........((.((.....--)).)).((((((((((((((((........)))))))))....((..((....))..))))))))) ( -30.60) >consensus CAAUGGGAAACUCGAAAUAAGCCAAGCAAACAGAAAUAAGCGGGGUGGAAAAAACAACGGCC__CCGUUGACUAAUUAGG_GGCAUUAGUCAACAGUGGCA___________________ ...(((....).))......((((................((..((.......))..)).......((((((((((........))))))))))..)))).................... (-16.83 = -17.00 + 0.17)

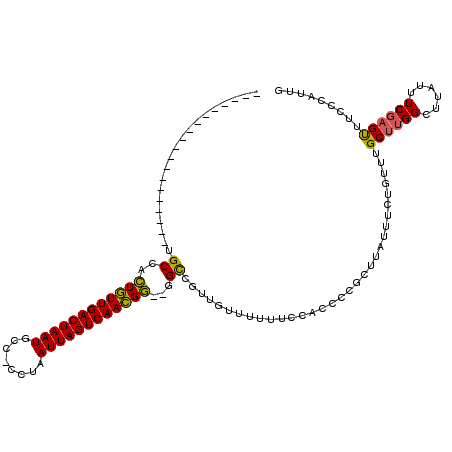
| Location | 14,814,729 – 14,814,827 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.35 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.07 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14814729 98 - 22407834 -------------------UGCCACUGUUGACUAAUGCC-CCUAAUUAGUCAACGG--GGCCGUUGUUUUUUCCACCUCGCUUAUUUCUGUUUGCUUGGCUUAUUUCGAGUUUCCCAUUG -------------------.(((.((((((((((((...-....))))))))))))--)))..............((..((..((....))..))..))..................... ( -24.60) >DroSec_CAF1 56218 98 - 1 -------------------UGCCACUGUUGACUAAUGCC-CCUAAUUAGUCAACGG--GGCCGUUGUUUUUUCCACCCCGGUUAUUUCUGUUUGCUUGGAUUAUUUCGAGUUUCCCAUUG -------------------.(((.((((((((((((...-....))))))))))))--))).................(((......)))...(((((((....)))))))......... ( -27.10) >DroSim_CAF1 56778 98 - 1 -------------------UGCCACUGUUGACUAAUGCC-CCUAAUUAGUCAACGG--GGCCGUUGUUUUUUCCACCCCGCUUAUUUCUGUUUGCUUGGCUUAUUUCGAGUUUCCCAUUG -------------------.((((..((((((((((...-....))))))))))((--((...............))))((..((....))..)).)))).................... ( -24.86) >DroEre_CAF1 52014 98 - 1 -------------------UGCCACUGUUGACUAAUGCC-CCUAAUUAGUCAACGG--GGCCAUUGUUUUUUCCACCCCGCUAAUUUCUGUUUGCUUGGCUUAUUUCGAGUUUCCCAUUG -------------------.((((..((((((((((...-....))))))))))((--((...............))))((.(((....))).)).)))).................... ( -24.96) >DroYak_CAF1 53628 98 - 1 -------------------UGCCACUGUUGACUAAUGCC-CCUAAUUAGUCAACGG--GGCCGUUGUUUUUUCCACCCCGCUUAUUUCUGUUUGCUUGGCUUAUUUCGAGUUUCCCAUUG -------------------.((((..((((((((((...-....))))))))))((--((...............))))((..((....))..)).)))).................... ( -24.86) >DroAna_CAF1 51749 117 - 1 CCGGGCCGGGACCGGACAGAGCCUGUUUUGACUAAUGCCUUCUAAUUAGUCAAAGCCCAGUGGCUG--UUUUUCACCCUCAUUGUUU-UUUCUGCUGGGUUCAUUUCGUGCUUCCCAUUG ..(((..(((..((((..((((((((((((((((((........)))))))))))).((((((..(--(.....))..))))))...-........))))))..))))..)))))).... ( -34.70) >consensus ___________________UGCCACUGUUGACUAAUGCC_CCUAAUUAGUCAACGG__GGCCGUUGUUUUUUCCACCCCGCUUAUUUCUGUUUGCUUGGCUUAUUUCGAGUUUCCCAUUG ....................((..((((((((((((........))))))))))))...))................................((((((......))))))......... (-19.56 = -19.07 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:50 2006