
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,809,524 – 14,809,719 |
| Length | 195 |
| Max. P | 0.999794 |

| Location | 14,809,524 – 14,809,623 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.07 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -28.16 |
| Energy contribution | -28.85 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
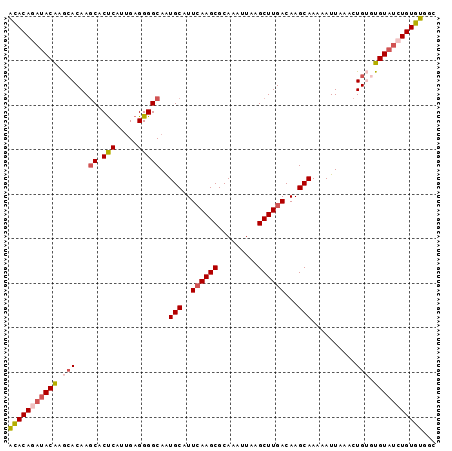
>2L_DroMel_CAF1 14809524 99 - 22407834 GAGUAGUCAGGUUGUUCUGGCUGU---UGUGAGUGUGGCAGUGCUUGUAUCUGUGUGUUUUGGAUUGCAUGCACGAAUAACAGGACACGGCAGGAAAUGCCA ..((.(((..(((((((..((..(---.......)..)).((((.((((((((.......)))).)))).)))))))))))..)))))((((.....)))). ( -33.60) >DroSec_CAF1 50943 102 - 1 GAGUAGUCAGGUUGUUCUGGCUGUGAGUGUGAGUGUGGCAGUGCUUGUAUCUGUGUGUUUUGGAUUGCAUGCACGAAUAAUAGGACACGGCAGGAAAUGCCA ..((.(((..(((((((..((..((........))..)).((((.((((((((.......)))).)))).)))))))))))..)))))((((.....)))). ( -31.60) >DroSim_CAF1 51604 102 - 1 GAGUAGUCAGGUUGUUCUGGCUGUGAGUGUGAGUGUGGCAGUGCUUGUAUCUGUGUGUUUUGGAUUGCAUGCACGAAUAACAGGACACGGCAGGAAAUGCCA ..((.(((..(((((((..((..((........))..)).((((.((((((((.......)))).)))).)))))))))))..)))))((((.....)))). ( -33.10) >DroYak_CAF1 47727 98 - 1 GAG-UGUCAGUUUGUUCUGGCUGU---UGUGAGUGUGGCAGUGCUUGUAUCUGUGUGUUUUGGAUUGCAUGCACGAAUAGCAGGACACGGCAGGAAAUGGCA ...-(((((.(((((..((.((((---(((..((((((((((.(.................).))))).)))))..)))))))..))..)))))...))))) ( -30.23) >consensus GAGUAGUCAGGUUGUUCUGGCUGU___UGUGAGUGUGGCAGUGCUUGUAUCUGUGUGUUUUGGAUUGCAUGCACGAAUAACAGGACACGGCAGGAAAUGCCA .....(((..(((((((..((..((........))..)).((((.((((((((.......)))).)))).)))))))))))..)))..((((.....)))). (-28.16 = -28.85 + 0.69)

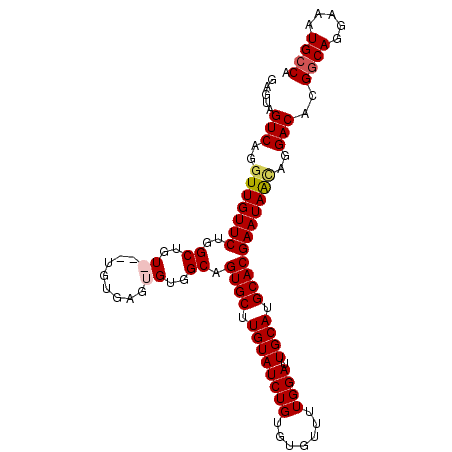

| Location | 14,809,623 – 14,809,719 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.15 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -25.48 |
| Energy contribution | -26.64 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14809623 96 + 22407834 ACACAAAUACAAGCACAAGCACUCAUUGAGGGGCAAUGCAUUCAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUGUAUGUAGCUGUGUGGC (((((..((((.(((...((.(((.....)))))..(((..((((((.........))))))...)))..........))).))))..)))))... ( -25.50) >DroSec_CAF1 51045 96 + 1 ACACAGAUACGAGCACAAGCACUCAUUGAGGGGCAAUGCAUUCAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUGUGUGUAUCUGUGUGGC ((((((((((..(((((.((.(((.....)))))..(((..((((((.........))))))...)))..........)))))))))))))))... ( -33.40) >DroSim_CAF1 51706 96 + 1 ACACAGAUACAAGCACAAGCACUCAUUGAGGGGCAAUGCAUUCAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUGUGUGUAUCUGUGUGGC ((((((((((..(((((.((.(((.....)))))..(((..((((((.........))))))...)))..........)))))))))))))))... ( -33.40) >DroYak_CAF1 47825 96 + 1 GCACAGAUACACACACAAGCACUCAUUGAGGGGCAAUGCAUUCAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUGAGUGUAUCUGUGUGGC ((((((((((((......((.(((.....)))))..(((..((((((.........))))))...))).............))))))))))))... ( -33.90) >DroAna_CAF1 36914 94 + 1 AUACAAACACAGAGACAAACACCCAUCGAGGGGCAGUGCAUUGAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUG--UGUAUCUGUGUAGG ......(((((((.((.....(((.....)))(((((((......)).........(((.....))).........))))--))).)))))))... ( -24.20) >consensus ACACAGAUACAAGCACAAGCACUCAUUGAGGGGCAAUGCAUUCAAGCGCAAAUUAAGCUUGACAAGCAAAAAUUAAACUGUGUGUAUCUGUGUGGC (((((((((((.(((...((.(((.....)))))..(((..((((((.........))))))...)))..........))).)))))))))))... (-25.48 = -26.64 + 1.16)

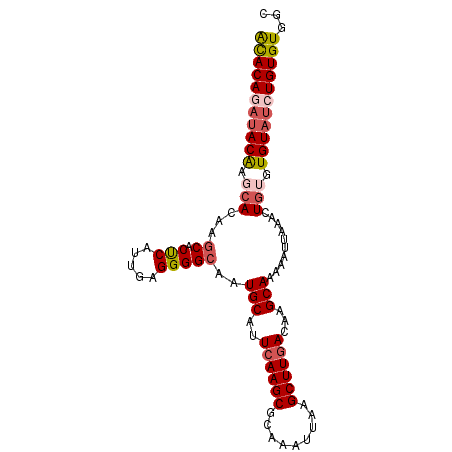
| Location | 14,809,623 – 14,809,719 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.15 |
| Mean single sequence MFE | -27.06 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14809623 96 - 22407834 GCCACACAGCUACAUACAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUGAAUGCAUUGCCCCUCAAUGAGUGCUUGUGCUUGUAUUUGUGU ...((((((.(((((((((.........(((...((((((.........))))))..)))..((..(((...))).)))))))..)))).)))))) ( -22.30) >DroSec_CAF1 51045 96 - 1 GCCACACAGAUACACACAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUGAAUGCAUUGCCCCUCAAUGAGUGCUUGUGCUCGUAUCUGUGU ...((((((((((...............(((...((((((.........))))))..)))............(((((....))))))))))))))) ( -29.70) >DroSim_CAF1 51706 96 - 1 GCCACACAGAUACACACAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUGAAUGCAUUGCCCCUCAAUGAGUGCUUGUGCUUGUAUCUGUGU ...((((((((((((((((.........(((...((((((.........))))))..)))..((..(((...))).)))))))..))))))))))) ( -30.60) >DroYak_CAF1 47825 96 - 1 GCCACACAGAUACACUCAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUGAAUGCAUUGCCCCUCAAUGAGUGCUUGUGUGUGUAUCUGUGC ....(((((((((((.((..........(((...((((((.........))))))..)))..((..(((...))).))...)).))))))))))). ( -30.70) >DroAna_CAF1 36914 94 - 1 CCUACACAGAUACA--CAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUCAAUGCACUGCCCCUCGAUGGGUGUUUGUCUCUGUGUUUGUAU ...(((((((.(((--............((((((....((.......))....))).)))..((((......))))...))).)))))))...... ( -22.00) >consensus GCCACACAGAUACACACAGUUUAAUUUUUGCUUGUCAAGCUUAAUUUGCGCUUGAAUGCAUUGCCCCUCAAUGAGUGCUUGUGCUUGUAUCUGUGU ...((((((((((((((((.........(((...((((((.........))))))..)))..((..(((...))).)))))))..))))))))))) (-22.84 = -23.60 + 0.76)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:46 2006