| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,799,284 – 14,799,400 |
| Length | 116 |
| Max. P | 0.713613 |

| Location | 14,799,284 – 14,799,400 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -14.11 |
| Energy contribution | -16.92 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
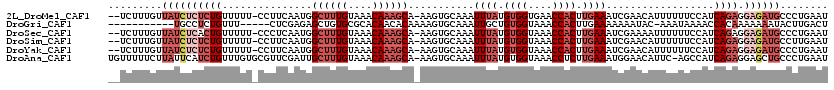
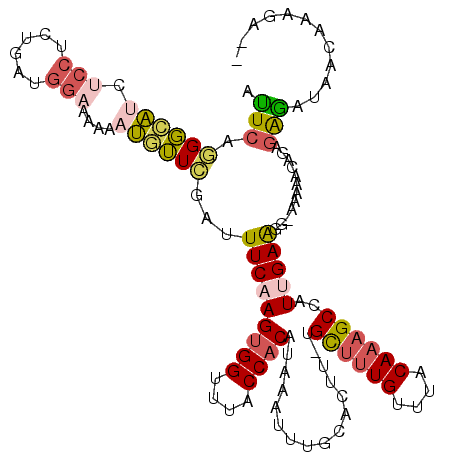
>2L_DroMel_CAF1 14799284 116 + 22407834 --UCUUUGUUAUCUCUCUGUUUUU-CCUUCAAUGGCUUUGUAAACAAAGCA-AAGUGCAAAUUUAUGUGGUGAACCACUUGAAAUCGAACAUUUUUUCCAUCAGAGGAGAUGCCCUGAAU --...........((...((.(((-((((..(((((((((....))))))(-(((((...((((..((((....))))...))))....))))))...)))..))))))).))...)).. ( -28.50) >DroGri_CAF1 50253 103 + 1 -----------UGCCUCUGUUU-----CUCGAGAGCUGUGCGCACAACACAAAAGUGCAAAUUGCUGUGGUAAACCACUUGAAAAAAAUAC-AAAUAAAACCACAAAAAAAUACUUGACU -----------...........-----.((((((((..(((((...........)))))....)))((((....)))).............-.....................))))).. ( -17.90) >DroSec_CAF1 45928 116 + 1 --UCUUUGUUAUCUCACUGUUUUU-CCCUCAAUGGCUUUGUAAACAAAGCA-AAGUGCAAAUUUAUGUGGUAAACCACUUGAAAUCGAAAAUUUUUUCCAUCAGAGGAGAUGCCCUGAAU --...........(((..((...(-(((((.(((((((((....)))))).-.........((((.((((....)))).))))...............)))..)))).)).))..))).. ( -23.90) >DroSim_CAF1 46488 116 + 1 --UCUUUGUUAUCUCUCUGUUUUU-CCUUCAAUGGCUUUGUAAACAAAGCA-AAGUGCAAAUUUAUGUGGUAAACCACUUGAAAUCGAACAUUUUUUCCAUCAGAGGAGAUGCCUUGAAU --...........((...((.(((-((((..(((((((((....))))))(-(((((...((((..((((....))))...))))....))))))...)))..))))))).))...)).. ( -27.00) >DroYak_CAF1 41849 116 + 1 --UCUUUGUUAUCUCUCUGUUUUU-CCUUCAAUGGCUUUGUAAACAAAGCA-AAGUGCAAAUUUAUGUGGUAAACCACUUGAAAUCGAACAUUUUUUCCAUCAGAGGAGAUGCCCUGAAU --...........((...((.(((-((((..(((((((((....))))))(-(((((...((((..((((....))))...))))....))))))...)))..))))))).))...)).. ( -27.00) >DroAna_CAF1 33901 118 + 1 UGUUUUUCUUAUUCAUCUGUUUGUGCGUUCGAUUGCUUUGUAAACAAAGCA-AAGUGCAAAUUUAUGUGGUAAACCUCUUGAAAUGGAACAUUC-AGCCAUCAGAGGAGCUGCCCUGAAU .....(((.....(((..(((((..(......((((((((....)))))))-).)..)))))..))).((((..(((((.((..(((.......-..))))))))))...))))..))). ( -28.80) >consensus __UCUUUGUUAUCUCUCUGUUUUU_CCUUCAAUGGCUUUGUAAACAAAGCA_AAGUGCAAAUUUAUGUGGUAAACCACUUGAAAUCGAACAUUUUUUCCAUCAGAGGAGAUGCCCUGAAU .........((((((((((...............((((((....))))))...........((((.((((....)))).))))..................)))).))))))........ (-14.11 = -16.92 + 2.81)
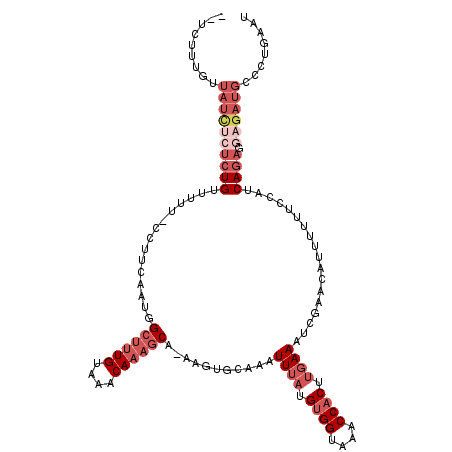
| Location | 14,799,284 – 14,799,400 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -15.83 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14799284 116 - 22407834 AUUCAGGGCAUCUCCUCUGAUGGAAAAAAUGUUCGAUUUCAAGUGGUUCACCACAUAAAUUUGCACUU-UGCUUUGUUUACAAAGCCAUUGAAGG-AAAAACAGAGAGAUAACAAAGA-- ......(..(((((.((((...(((......)))..((((((((((....))))..............-.((((((....))))))..)))))).-.....)))))))))..).....-- ( -27.20) >DroGri_CAF1 50253 103 - 1 AGUCAAGUAUUUUUUUGUGGUUUUAUUU-GUAUUUUUUUCAAGUGGUUUACCACAGCAAUUUGCACUUUUGUGUUGUGCGCACAGCUCUCGAG-----AAACAGAGGCA----------- .(((..(((..((.(((((((.((((((-(.........)))))))...))))))).))..))).(((..(.(((((....))))).)..)))-----.......))).----------- ( -25.70) >DroSec_CAF1 45928 116 - 1 AUUCAGGGCAUCUCCUCUGAUGGAAAAAAUUUUCGAUUUCAAGUGGUUUACCACAUAAAUUUGCACUU-UGCUUUGUUUACAAAGCCAUUGAGGG-AAAAACAGUGAGAUAACAAAGA-- ......(..(((((..(((...((((....))))..((((((((((....))))..............-.((((((....))))))..)))))).-.....))).)))))..).....-- ( -25.60) >DroSim_CAF1 46488 116 - 1 AUUCAAGGCAUCUCCUCUGAUGGAAAAAAUGUUCGAUUUCAAGUGGUUUACCACAUAAAUUUGCACUU-UGCUUUGUUUACAAAGCCAUUGAAGG-AAAAACAGAGAGAUAACAAAGA-- ......(..(((((.((((...(((......)))..((((((((((....))))..............-.((((((....))))))..)))))).-.....)))))))))..).....-- ( -27.40) >DroYak_CAF1 41849 116 - 1 AUUCAGGGCAUCUCCUCUGAUGGAAAAAAUGUUCGAUUUCAAGUGGUUUACCACAUAAAUUUGCACUU-UGCUUUGUUUACAAAGCCAUUGAAGG-AAAAACAGAGAGAUAACAAAGA-- ......(..(((((.((((...(((......)))..((((((((((....))))..............-.((((((....))))))..)))))).-.....)))))))))..).....-- ( -27.20) >DroAna_CAF1 33901 118 - 1 AUUCAGGGCAGCUCCUCUGAUGGCU-GAAUGUUCCAUUUCAAGAGGUUUACCACAUAAAUUUGCACUU-UGCUUUGUUUACAAAGCAAUCGAACGCACAAACAGAUGAAUAAGAAAAACA (((((((......((((((((((..-.......)))))...)))))....)).........(((...(-(((((((....))))))))......)))........))))).......... ( -24.30) >consensus AUUCAGGGCAUCUCCUCUGAUGGAAAAAAUGUUCGAUUUCAAGUGGUUUACCACAUAAAUUUGCACUU_UGCUUUGUUUACAAAGCCAUUGAAGG_AAAAACAGAGAGAUAACAAAGA__ .(((.((((((.(((......)))....))))))...(((((((((....))))................((((((....))))))..)))))............)))............ (-15.83 = -16.20 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:42 2006