
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,796,805 – 14,796,953 |
| Length | 148 |
| Max. P | 0.838453 |

| Location | 14,796,805 – 14,796,916 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
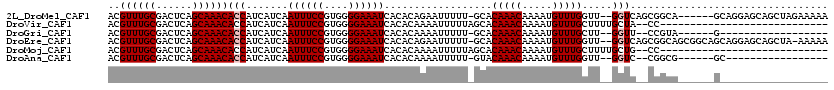
>2L_DroMel_CAF1 14796805 111 - 22407834 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAUUUUU-GCACAAACAAAAUGUUUGGUU--GGUCAGCGGCA------GCAGGAGCAGCUAGAAAAA ..((((((......))))))................(((.((((((........))))))-.)))..........(((((((--(.((.((....------))..)).)))))))).... ( -27.40) >DroVir_CAF1 58051 90 - 1 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUUAGCACAAACAAAAUGUUUGCUUUUGCUA--CC---------------------------- ..((((((......))))))..........((((((....)))))).............((((((((((.....)))))....)))))--..---------------------------- ( -20.10) >DroGri_CAF1 48021 91 - 1 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU-GCACAAACAAAAUGUUUGCUU--GGUU--CCGUA------G------------------ ..((((((......)))))).((((...........))))((((.(((..((((..((((-(......)))))..))))..)--))))--))...------.------------------ ( -20.50) >DroEre_CAF1 36307 116 - 1 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAUUUUU-GCACAAACAAAAUGUUUGGUU--GGUCAGCGGCAGCGGCAGCAGGAGCAGCUA-AAAAA ..((((((......))))))................(((.((((((........))))))-.)))..........(((((((--(.((.((.((....)).))..)).))))))-))... ( -29.50) >DroMoj_CAF1 58384 90 - 1 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUUAGCACAAACAAAAUGUUUGCUUUUGCUG--CC---------------------------- ..((((((......))))))..........((((((....)))))).............((((((((((.....)))))....)))))--..---------------------------- ( -19.80) >DroAna_CAF1 31363 92 - 1 ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU-GUACAAACAAAAUGUUUGGUU--GGUC--CGGCG------GC----------------- ..((((((......))))))..............((((.(((.(((((.(((....((((-((....))))))))).)))))--..))--).)))------).----------------- ( -26.70) >consensus ACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU_GCACAAACAAAAUGUUUGCUU__GGUC__CCGCA______G__________________ ..((((((......))))))(((.......((((((....))))))..................(((((.....))))).....)))................................. (-16.98 = -17.32 + 0.33)

| Location | 14,796,837 – 14,796,953 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.73 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -17.24 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
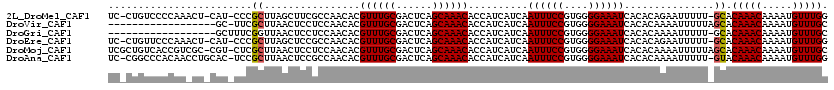
>2L_DroMel_CAF1 14796837 116 - 22407834 UC-CUGUCCCCAAACU-CAU-CCCGCUUAGCUUCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAUUUUU-GCACAAACAAAAUGUUUGG ..-......((((((.-..(-(((((...((...))......((((((......))))))................))))))..............((((-(......))))).)))))) ( -23.60) >DroVir_CAF1 58061 101 - 1 ------------------GC-UUCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUUAGCACAAACAAAAUGUUUGC ------------------..-...(((...............((((((......))))))..........((((((....))))))..............))).(((((.....))))). ( -18.70) >DroGri_CAF1 48033 101 - 1 ------------------GCUUUCGGUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU-GCACAAACAAAAUGUUUGC ------------------((....((......))(((.....((((((......)))))).((((...........))))))).................-)).(((((.....))))). ( -19.40) >DroEre_CAF1 36344 116 - 1 UC-CUGUUCCCAAACU-CAU-CCCGCUUAGCUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAGAAUUUUU-GCACAAACAAAAUGUUUGG ..-......((((((.-..(-(((((...((...))......((((((......))))))................))))))..............((((-(......))))).)))))) ( -23.60) >DroMoj_CAF1 58394 118 - 1 UCGCUGUCACCGUCGC-CGU-CUCGCUUAACUCCUCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUUAGCACAAACAAAAUGUUUGC ..((((........((-...-...))........(((.....((((((......)))))).((((...........)))))))................)))).(((((.....))))). ( -21.40) >DroAna_CAF1 31376 117 - 1 UC-CGGCCCACAACCUGCAC-UCCGCUUAACUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU-GUACAAACAAAAUGUUUGG ..-...(((((.....((..-...))................((((((......))))))................))))).........((((..((((-((....))))))..)))). ( -24.00) >consensus UC_CUGUCCCCAA_CU_CAC_CCCGCUUAACUCCGCCAACACGUUUGCGACUCAGCAAACACCAUCAUCAAUUUCCGUGGGGAAAUCACACAAAAUUUUU_GCACAAACAAAAUGUUUGC ........................((................((((((......))))))..........((((((....))))))...............)).(((((.....))))). (-17.24 = -17.27 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:37 2006