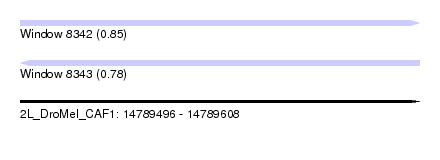
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,789,496 – 14,789,608 |
| Length | 112 |
| Max. P | 0.846967 |

| Location | 14,789,496 – 14,789,608 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -11.71 |
| Energy contribution | -14.06 |
| Covariance contribution | 2.35 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14789496 112 + 22407834 CCUAACAUUGGGACGCCGAGCACAAGCUCAGCGGCAGGACUCAAAGCCCUUCUA--GUUAAGCCUGGA-AAUACAUUUGAAAUUUAUCAAAAAUAAAUUCCUCGUCUGCCAUUUU ((((....))))..((.((((....)))).))(((((.(((((((....(((((--(......)))))-).....)))))(((((((.....)))))))....)))))))..... ( -29.90) >DroSec_CAF1 35984 112 + 1 CCUUACAUCGGGACGCAGAGCACAAGCUCAGCGGCAGGACUCAAAGCUCUUCUA--GUUAAGCUUUGA-AAUACAUUUGAAAUUUAUUAAAAAUAUAUUCCUCGUCUGCCAUUUU (((......)))..((.((((....)))).))(((((.((((((((((......--....))))))))-......(((((......)))))............)))))))..... ( -27.50) >DroSim_CAF1 36580 112 + 1 CCUUACAUCGGGACGCAGAGCACAAGCUCAGCGGUAUGACUCAAAGCUCUUCUA--GUUAAGCUUUGA-AAUACAUUUGAAAUUUAUUAAAAAUAUAUUCCUCGUCUGCCAUUUU (((......)))..((.((((....)))).))((((.(((((((((((......--....))))))))-......(((((......)))))............)))))))..... ( -25.50) >DroEre_CAF1 28939 112 + 1 CCUUACAUAGGAACACACAUCACAAGCUCAACGGCAGGACUCAAAGCUCUCCUA--GUUAAGCUUUGA-AAUACAUUUAAAAUUUAUUAAAAGUAAAUUCCUCGUCUGCCAUUUU (((.....))).....................(((((.((((((((((......--....))))))))-..(((.(((((......))))).)))........)))))))..... ( -22.20) >DroAna_CAF1 25536 97 + 1 CCUUACAUAGU-----------------GAGCAACAGGACUCGAAACUCUUCUUUCCUCAAGCUUUGAAAAUUCAUUUGAAAUUUAUUAAAAAACAAAGCCGC-UCUGCCUUUUC ...........-----------------((((...((((...(((....)))..))))...((((((....(((....))).(((....)))..)))))).))-))......... ( -14.20) >consensus CCUUACAUAGGGACGCAGAGCACAAGCUCAGCGGCAGGACUCAAAGCUCUUCUA__GUUAAGCUUUGA_AAUACAUUUGAAAUUUAUUAAAAAUAAAUUCCUCGUCUGCCAUUUU ..............((.((((....)))).))((((.(((((((((((............)))))))).......(((((......)))))............)))))))..... (-11.71 = -14.06 + 2.35)

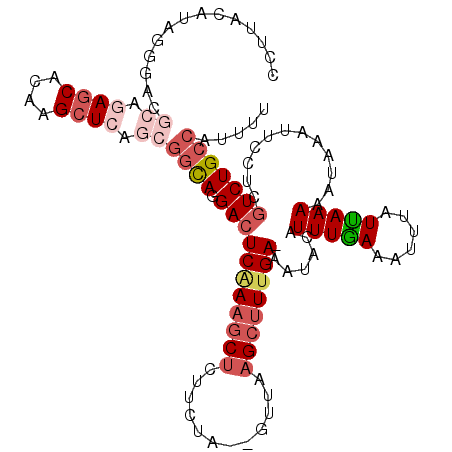

| Location | 14,789,496 – 14,789,608 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -29.06 |
| Consensus MFE | -18.99 |
| Energy contribution | -20.98 |
| Covariance contribution | 1.99 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
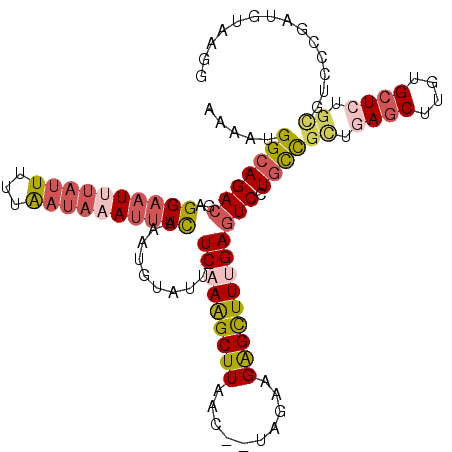
>2L_DroMel_CAF1 14789496 112 - 22407834 AAAAUGGCAGACGAGGAAUUUAUUUUUGAUAAAUUUCAAAUGUAUU-UCCAGGCUUAAC--UAGAAGGGCUUUGAGUCCUGCCGCUGAGCUUGUGCUCGGCGUCCCAAUGUUAGG .....(((((((..((((..(((.(((((......))))).))).)-)))((((((...--.....))))))...)).)))))(((((((....))))))).............. ( -36.20) >DroSec_CAF1 35984 112 - 1 AAAAUGGCAGACGAGGAAUAUAUUUUUAAUAAAUUUCAAAUGUAUU-UCAAAGCUUAAC--UAGAAGAGCUUUGAGUCCUGCCGCUGAGCUUGUGCUCUGCGUCCCGAUGUAAGG .....(((((((..(((((((((((..((....))..)))))))))-))(((((((...--.....)))))))..)).)))))((.((((....)))).)).............. ( -33.10) >DroSim_CAF1 36580 112 - 1 AAAAUGGCAGACGAGGAAUAUAUUUUUAAUAAAUUUCAAAUGUAUU-UCAAAGCUUAAC--UAGAAGAGCUUUGAGUCAUACCGCUGAGCUUGUGCUCUGCGUCCCGAUGUAAGG ......(((..((.(((......................(((...(-(((((((((...--.....)))))))))).)))..(((.((((....)))).)))))))).))).... ( -29.90) >DroEre_CAF1 28939 112 - 1 AAAAUGGCAGACGAGGAAUUUACUUUUAAUAAAUUUUAAAUGUAUU-UCAAAGCUUAAC--UAGGAGAGCUUUGAGUCCUGCCGUUGAGCUUGUGAUGUGUGUUCCUAUGUAAGG ..((((((((((........(((.(((((......))))).)))..-(((((((((...--.....))))))))))).))))))))((((...........)))).......... ( -27.90) >DroAna_CAF1 25536 97 - 1 GAAAAGGCAGA-GCGGCUUUGUUUUUUAAUAAAUUUCAAAUGAAUUUUCAAAGCUUGAGGAAAGAAGAGUUUCGAGUCCUGUUGCUC-----------------ACUAUGUAAGG .........((-(((((((((...((((............))))....)))))))..((((..(((....)))...))))...))))-----------------........... ( -18.20) >consensus AAAAUGGCAGACGAGGAAUUUAUUUUUAAUAAAUUUCAAAUGUAUU_UCAAAGCUUAAC__UAGAAGAGCUUUGAGUCCUGCCGCUGAGCUUGUGCUCUGCGUCCCGAUGUAAGG .....(((((((..((((((((((...))))))))))..........(((((((((..........)))))))))))).))))((.((((....)))).)).............. (-18.99 = -20.98 + 1.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:30 2006