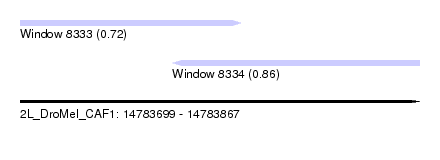
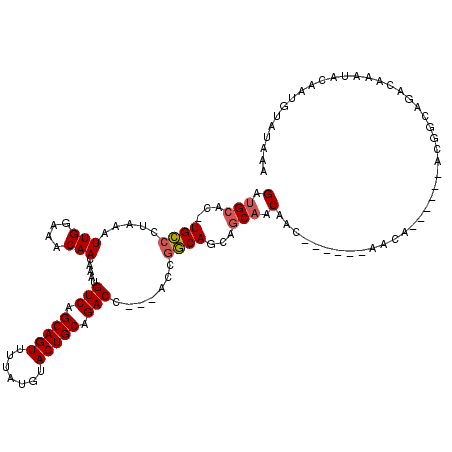
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,783,699 – 14,783,867 |
| Length | 168 |
| Max. P | 0.864238 |

| Location | 14,783,699 – 14,783,792 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.74 |
| Mean single sequence MFE | -13.95 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14783699 93 + 22407834 CACACACAUACA---------CAUACACUCGCACAGGCACAUCCUUGUGCAUUCAUCAUCAGCCACAUCAUCAUUUAUACAUUGUAUUUGUCUGCCGUUGUU ......((.(((---------.(((((...(((((((......)))))))...........(......).............))))).))).))........ ( -13.00) >DroSec_CAF1 30058 87 + 1 CACACACA---------------CACACUCGCACAGGCACAUCCUUGUGCAUUCAUCAUCAGCCACAUCAUCAUUUAUACAUUGUAUUUGUCUGACGUUGUU .....(((---------------.((....(((((((......)))))))........((((.((.((((............)).)).)).)))).))))). ( -13.80) >DroSim_CAF1 28091 87 + 1 CACACACA---------------CACACUCGCACAGGCACAUCCUUGUGCAUUCAUCAUCAGCCACAUCAUCAUUUAUACAUUGUAUUUGUCUGCCGUUGUU ........---------------.......(((((((......))))))).........((((..((....((..(((.....)))..))..))..)))).. ( -12.60) >DroEre_CAF1 23215 102 + 1 CACACACACACACACAACACCCAUACGCUGGCACUGGCACAUCCUUGUGCAUUCAUCAUCAGCCACAUCAUCAUUUAUACAUUGUAUUUGUCUGCUGUUGUU .............((((((..((.(((((((.....(((((....)))))........))))).(((...............)))....)).)).)))))). ( -16.38) >consensus CACACACA_______________CACACUCGCACAGGCACAUCCUUGUGCAUUCAUCAUCAGCCACAUCAUCAUUUAUACAUUGUAUUUGUCUGCCGUUGUU ..............................(((((((......))))))).........((((..((....((..(((.....)))..))..))..)))).. (-11.40 = -11.65 + 0.25)

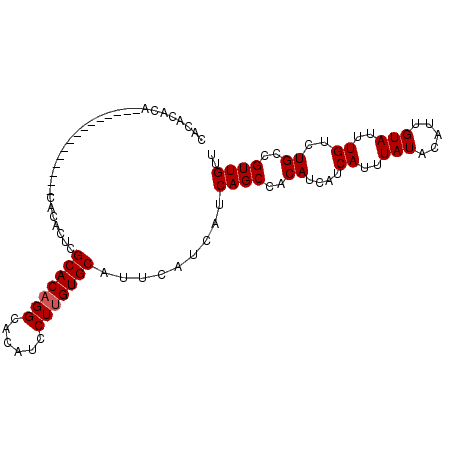
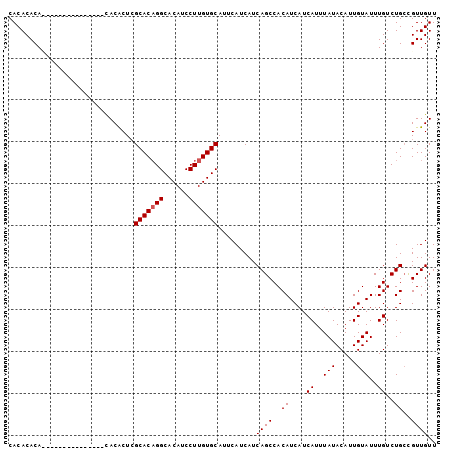
| Location | 14,783,763 – 14,783,867 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.62 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.37 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14783763 104 - 22407834 GAUGCAC-UGCCCUAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCGGACC---ACUGGCAGCAGCAGCAAC------AACA------ACGGCAGACAAAUACAAUGUAUAAA ..(((.(-((((.....(((....))).....(((.(((((.......))))).))).---...)))))..)))((...------....------...)).................... ( -24.10) >DroSec_CAF1 30116 107 - 1 GAUGCAC-UGCCCUAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACCACCACCGGCAGCAGCAACAAC------AACA------ACGUCAGACAAAUACAAUGUAUAAA ..(((.(-((((.....(((....))).....(((.(((((.......))))).))).......)))))..))).....------....------......................... ( -23.50) >DroSim_CAF1 28149 107 - 1 GAUGCAC-UGCCCUAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACCACCACCGGCAGCAGCAACAAC------AACA------ACGGCAGACAAAUACAAUGUAUAAA ..(((.(-((((.....(((....))).....(((.(((((.......))))).))).......)))))..))).....------....------......................... ( -23.50) >DroEre_CAF1 23288 104 - 1 GAUGCAC-UGCCCCAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACC---ACCGGCAGCAGCACCAAC------AACA------ACAGCAGACAAAUACAAUGUAUAAA (.(((.(-((((.....(((....))).....(((.(((((.......))))).))).---...)))))..))).)...------....------......................... ( -23.50) >DroYak_CAF1 27299 104 - 1 GAUGCAC-UGCCCUAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACC---ACCGGCAGCAGCAACAAC------AACA------GCAGCAGACAAAUACAAUGUAUAAA ..(((.(-((((.....(((....))).....(((.(((((.......))))).))).---...)))))..((......------....------)).)))................... ( -24.10) >DroAna_CAF1 20082 117 - 1 GCCGCCCGUGUCCCAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACC---GUCAACAGAAACAACAACAGCAACAACAAAAGCUACGCCAGACAAAUACAAUGUAUAAA .......((((......(((....)))....((((.(((((.......))))).....---..................(((..........)))......)))).)))).......... ( -17.20) >consensus GAUGCAC_UGCCCUAAAUUGGAAACAACAAAUGUCAGCAGUUUUAUGUACUGCAGACC___ACCGGCAGCAGCAACAAC______AACA______ACGGCAGACAAAUACAAUGUAUAAA (.(((...((((.....(((....))).....(((.(((((.......))))).))).......))))...))).)............................................ (-19.14 = -19.37 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:21 2006