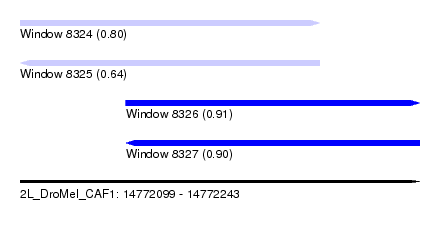
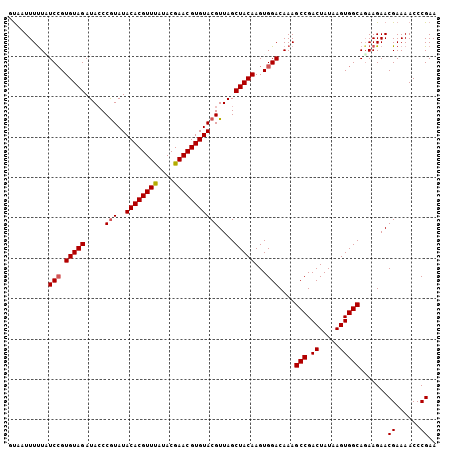
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,772,099 – 14,772,243 |
| Length | 144 |
| Max. P | 0.911092 |

| Location | 14,772,099 – 14,772,207 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -24.36 |
| Consensus MFE | -17.38 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14772099 108 + 22407834 CUAU-UUUUCUU-AUACCAUAAAAACUUAUAAAGAAGACUGUAAUUUUUAUCCGUGUAGAUACCCAUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAA ....-.....((-((((.(((((((.(((((........))))))))))))..))))))....((((.(((((((((....))))))))).((.....))..)))).... ( -19.50) >DroSec_CAF1 18629 108 + 1 CUAU-UUUUCUU-UUAUCAUAAAACCUUAUAAGGAAGUCAGUAAUUUUUAUCCGUGUAGAUAGCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAA ((..-(((((((-.((...........)).)))))))..)).........(((.(((((.((((....(((((((((....))))))))).)))).)))))...)))... ( -22.60) >DroSim_CAF1 16353 109 + 1 CUAU-UUUUUUUAUUAUCAUAAAACCUUAUAGAGGAAUCUAUAAUUUUUAUCCGUGUAGAUACCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAA ....-..............(((((..(((((((....))))))).)))))(((.(((((.....(((..((((((((....)))))))))))....)))))...)))... ( -23.70) >DroEre_CAF1 12788 108 + 1 CAAUCUUUCCUC-AUUUCAUAAGAAGUUAGAA-GGGGACUUCAAUUUCUCUCCGUGUAGAUACCCGUAUACACGUUCAUACGAACGUGUACGUUAGCUACAAGUGGACAA .......(((.(-........(((((((.(((-(....))))))))))).....(((((.....(((..((((((((....)))))))))))....))))).).)))... ( -32.70) >DroYak_CAF1 15679 108 + 1 CUAU-UUCUUUU-AUUUCACAAAACGCUAAAAUGAAGACUGUAAUUUUUUUCGGUGUAGAUACCCGUAUACACGUUCAUACGAACGUGUACGUUAGCUACAAGUGGACAA ....-.......-..(((((.....(((((.....................(((.((....)))))..(((((((((....)))))))))..))))).....)))))... ( -23.30) >consensus CUAU_UUUUCUU_AUAUCAUAAAACCUUAUAAAGAAGACUGUAAUUUUUAUCCGUGUAGAUACCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAA ................((((........(((((((.........)))))))...(((((.....(((..((((((((....)))))))))))....))))).)))).... (-17.38 = -16.62 + -0.76)

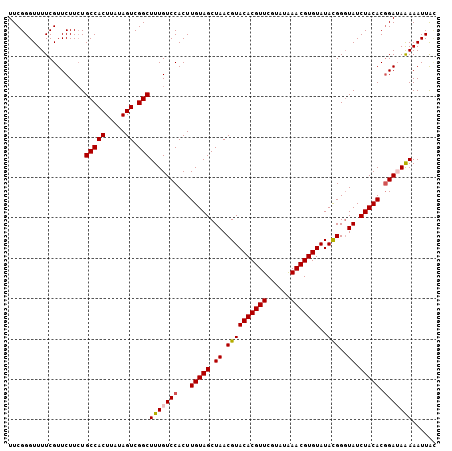
| Location | 14,772,099 – 14,772,207 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -18.06 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14772099 108 - 22407834 UUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUAUGGGUAUCUACACGGAUAAAAAUUACAGUCUUCUUUAUAAGUUUUUAUGGUAU-AAGAAAA-AUAG ((((((...(((((((...(((((((((......)))))))))...))...))))).)))))).........((((..((((((....))))))...-))))...-.... ( -21.60) >DroSec_CAF1 18629 108 - 1 UUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGCUAUCUACACGGAUAAAAAUUACUGACUUCCUUAUAAGGUUUUAUGAUAA-AAGAAAA-AUAG ((((((....((((((...(((((((((......)))))))))..))))))......))))))..((((..(((((.......)))))...))))..-.......-.... ( -22.60) >DroSim_CAF1 16353 109 - 1 UUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGGUAUCUACACGGAUAAAAAUUAUAGAUUCCUCUAUAAGGUUUUAUGAUAAUAAAAAAA-AUAG ((((((...(((((.((.((((((((((......)))))))..)))..)).))))).))))))..((((((((..(((.....))).))))))))..........-.... ( -26.40) >DroEre_CAF1 12788 108 - 1 UUGUCCACUUGUAGCUAACGUACACGUUCGUAUGAACGUGUAUACGGGUAUCUACACGGAGAGAAAUUGAAGUCCCC-UUCUAACUUCUUAUGAAAU-GAGGAAAGAUUG ...(((.(.(((((.((.(((((((((((....))))))))..)))..)).))))).)..(((((.((((((....)-))).)).))))).......-..)))....... ( -27.10) >DroYak_CAF1 15679 108 - 1 UUGUCCACUUGUAGCUAACGUACACGUUCGUAUGAACGUGUAUACGGGUAUCUACACCGAAAAAAAUUACAGUCUUCAUUUUAGCGUUUUGUGAAAU-AAAAGAA-AUAG .....(((.....(((((.((((((((((....)))))))))).(((((....)).))).....................))))).....)))....-.......-.... ( -25.30) >consensus UUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGGUAUCUACACGGAUAAAAAUUACAGUCUUCUUUAUAAGGUUUUAUGAUAU_AAGAAAA_AUAG ((((((...(((((.((.((((((((((......)))))))..)))..)).))))).))))))............................................... (-18.06 = -18.34 + 0.28)

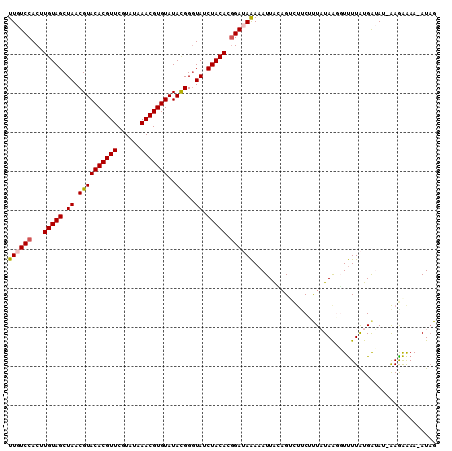
| Location | 14,772,137 – 14,772,243 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.911092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14772137 106 + 22407834 GUAAUUUUUAUCCGUGUAGAUACCCAUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAGAAGAACGAAAACCCGAA ....(((((.(((.(((((......(..(((((((((....)))))))))..)...)))))...)))....(((.((.....))))).))))).((......)).. ( -23.00) >DroSec_CAF1 18667 106 + 1 GUAAUUUUUAUCCGUGUAGAUAGCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAUAAGAACGAAAACCCGAA ....(((((((((.(((((.((((....(((((((((....))))))))).)))).)))))...)))....(((.((.....))))).))))))((......)).. ( -26.40) >DroSim_CAF1 16392 106 + 1 AUAAUUUUUAUCCGUGUAGAUACCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAGAAGAACGAAAACCCGAA ....(((((.(((.(((((.....(((..((((((((....)))))))))))....)))))...)))....(((.((.....))))).))))).((......)).. ( -24.30) >DroEre_CAF1 12826 106 + 1 UCAAUUUCUCUCCGUGUAGAUACCCGUAUACACGUUCAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAGAAGAACGAAAACCCGAA ....(((((.(((.(((((.....(((..((((((((....)))))))))))....)))))...)))....(((.((.....))))))))))..((......)).. ( -27.70) >DroYak_CAF1 15717 106 + 1 GUAAUUUUUUUCGGUGUAGAUACCCGUAUACACGUUCAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAGAAGAACGAAAACCCGAA .........(((((.((...........(((((((((....)))))))))((((.(((((.(((((......)).)))....))))).....))))...))))))) ( -28.20) >consensus GUAAUUUUUAUCCGUGUAGAUACCCGUAUACACGUUUAUACGAACGUGUACGUUAGCUACAAGUGGACAAAGCCGACUAUAAGUGGCAGAAGAACGAAAACCCGAA ..........(((.(((((.....(((..((((((((....)))))))))))....)))))...)))....(((.((.....))))).......((......)).. (-22.64 = -22.80 + 0.16)

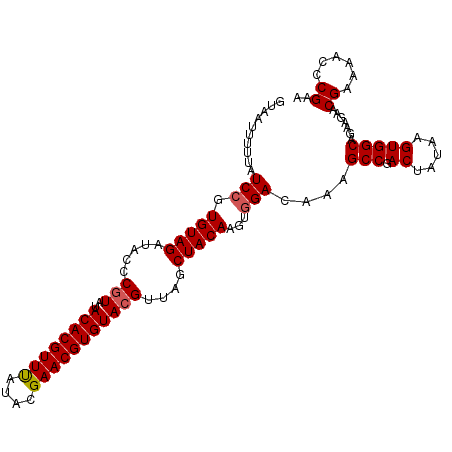
| Location | 14,772,137 – 14,772,243 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.85 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14772137 106 - 22407834 UUCGGGUUUUCGUUCUUCUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUAUGGGUAUCUACACGGAUAAAAAUUAC ...................(((((.....)).)))(((((((...(((((((...(((((((((......)))))))))...))...))))).)))))))...... ( -25.20) >DroSec_CAF1 18667 106 - 1 UUCGGGUUUUCGUUCUUAUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGCUAUCUACACGGAUAAAAAUUAC ...................(((((.....)).)))(((((((....((((((...(((((((((......)))))))))..))))))......)))))))...... ( -26.70) >DroSim_CAF1 16392 106 - 1 UUCGGGUUUUCGUUCUUCUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGGUAUCUACACGGAUAAAAAUUAU ...................(((((.....)).)))(((((((...(((((.((.((((((((((......)))))))..)))..)).))))).)))))))...... ( -25.70) >DroEre_CAF1 12826 106 - 1 UUCGGGUUUUCGUUCUUCUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUGAACGUGUAUACGGGUAUCUACACGGAGAGAAAUUGA .((((.(((((........(((((.....)).)))....(((...(((((.((.(((((((((((....))))))))..)))..)).))))).)))))))).)))) ( -29.50) >DroYak_CAF1 15717 106 - 1 UUCGGGUUUUCGUUCUUCUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUGAACGUGUAUACGGGUAUCUACACCGAAAAAAAUUAC (((((......(..(....(((((.....)).)))...)..)...(((((.((.(((((((((((....))))))))..)))..)).))))))))))......... ( -28.80) >consensus UUCGGGUUUUCGUUCUUCUGCCACUUAUAGUCGGCUUUGUCCACUUGUAGCUAACGUACACGUUCGUAUAAACGUGUAUACGGGUAUCUACACGGAUAAAAAUUAC ...................(((((.....)).)))(((((((...(((((.((.((((((((((......)))))))..)))..)).))))).)))))))...... (-23.64 = -23.92 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:14 2006