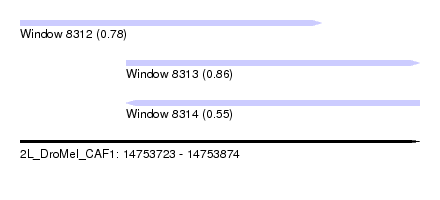
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,753,723 – 14,753,874 |
| Length | 151 |
| Max. P | 0.858727 |

| Location | 14,753,723 – 14,753,837 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.73 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -22.80 |
| Energy contribution | -21.12 |
| Covariance contribution | -1.68 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14753723 114 + 22407834 GAGCCUAAGCAGGUUCUUGGAUUUCCUCUCAUCUAGCCGUGGGGACAUUGAGAAUAUUCCGGGCUGCAU------UCAGCUUUUGGAAAAAGGACGGCAUAGUUGCCGUUGCAAUUUCUG ((((((....))))))...(((.(((((.(........).))))).))).((((..((((((((((...------.)))))..)))))....(((((((....))))))).....)))). ( -35.50) >DroSec_CAF1 17906 113 + 1 GAGCCUAAAAAGGUUCUUGGAUUUCCUCUCAUCUUGCCGUGGGGACAUUGAGAAUAUUCCGAGCUGCAU------GCAGCUU-UGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUG ((((((....))))))..((....))(((((...((((....)).)).)))))...((((((((((...------.)))).)-)))))....(((((((....))))))).......... ( -36.90) >DroSim_CAF1 17672 114 + 1 GAGCCUAAGAAGGUUCUUGGAUUUCCUCUCAUCUUGCCGUGGGGACAUUGAGAAUAUUCCGAGCUGCAU------GCAGCUUUUGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUG ((((((....))))))..(((.....(((((...((((....)).)).)))))....)))((((((...------.))))))..(((((...(((((((....)))))))....))))). ( -36.40) >DroEre_CAF1 18492 113 + 1 GAGCCUAAGCAGGUUGUUCAAUU-CCGCUUAUCUGGCCGUGGGGACAUUGAAAAUAUAGCGAGUUGCAU------GCGGCUUUUGGUAAGAGGGCGGCAUAGUUGCUGUUGCAAUUUCUG .......((.(((((((......-((.((((((.((((((((.(((.(((.........)))))).).)------))))))...)))))).))((((((....)))))).))))))))). ( -33.50) >DroYak_CAF1 20028 112 + 1 GAGCCUAAGCAGGUUGUUGAAUU-CC-UUUAUCUGGCCGUGGGGACAUUGAAAACAUUCCGAGCUGCAU------GCAGCUCUUGGGGAGAGAAAGGCACAGUUGCUGUUGCAAUUUCUG .(((((....)))))........-((-(((.(((..(((((....)))............((((((...------.))))))..))..))).)))))...((((((....)))))).... ( -31.90) >DroAna_CAF1 37385 104 + 1 GAGCCUAU-----UUGU-------CC-UUUAGCUA--CUUGGGUGCAUUGAAAAUGUCCCCUGCCACCGAUAUUUGUGUCA-GUGGCAGGGGGAUGUGGCAGUUUCUGUUGCAAUUUCUG (.(((((.-----..((-------..-....))..--..))))).)...((((...(((((((((((.((((....)))).-))))))))))).((..((((...))))..)).)))).. ( -40.30) >consensus GAGCCUAAGCAGGUUCUUGGAUU_CCUCUCAUCUAGCCGUGGGGACAUUGAAAAUAUUCCGAGCUGCAU______GCAGCUUUUGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUG .(((((....)))))............(((((......)))))((.((((......(((((((((((........)))))))..))))....(((((((....))))))).)))).)).. (-22.80 = -21.12 + -1.68)


| Location | 14,753,763 – 14,753,874 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -23.88 |
| Energy contribution | -22.37 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.62 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14753763 111 + 22407834 GGGGACAUUGAGAAUAUUCCGGGCUGCAU------UCAGCUUUUGGAAAAAGGACGGCAUAGUUGCCGUUGCAAUUUCUGUUUUGAAAUUGGCUUAUUUACGUA-UUU--UUUAUGCGCC ((.(.(((.(((((((((((((((((...------.)))))..))))..((((((((((....))))))).(((((((......))))))).)))......)))-)))--)).)))).)) ( -35.30) >DroSec_CAF1 17946 110 + 1 GGGGACAUUGAGAAUAUUCCGAGCUGCAU------GCAGCUU-UGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUGUUUUGAAAUUGGCUUAUUUACGUA-UUU--UUUAUGCGAA (....)..((((....((((((((((...------.)))).)-)))))....(((((((....))))))).(((((((......))))))).))))....((((-(..--...))))).. ( -33.90) >DroSim_CAF1 17712 111 + 1 GGGGACAUUGAGAAUAUUCCGAGCUGCAU------GCAGCUUUUGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUGUUUUGAAAUUGGCUUAUUUACGUA-UUU--UUUAUGCGCC ((.(.(((.(((((((((((((((((...------.))))))..)))..((((((((((....))))))).(((((((......))))))).)))......)))-)))--)).)))).)) ( -35.40) >DroEre_CAF1 18531 113 + 1 GGGGACAUUGAAAAUAUAGCGAGUUGCAU------GCGGCUUUUGGUAAGAGGGCGGCAUAGUUGCUGUUGCAAUUUCUGUUUUGUAAUUGGCUUAUUUACGUA-UUUUUUUUCUGCGCC (.(((....(((((((((((.((((((((------((.((((((.....)))))).))))((((((....))))))........)))))).))).......)))-)))))..))).)... ( -31.91) >DroYak_CAF1 20066 111 + 1 GGGGACAUUGAAAACAUUCCGAGCUGCAU------GCAGCUCUUGGGGAGAGAAAGGCACAGUUGCUGUUGCAAUUUCUGUUUUGUAAUUGGCUUAUUUACGUA-UUU--UUUCUGCGCC ..(((...((....)).)))((((((...------.))))))..(.((((((((((((.(((((((....(((.....)))...))))))))))).........-)))--))))).)... ( -29.70) >DroAna_CAF1 37410 117 + 1 GGGUGCAUUGAAAAUGUCCCCUGCCACCGAUAUUUGUGUCA-GUGGCAGGGGGAUGUGGCAGUUUCUGUUGCAAUUUCUGUUUUGAAAUUGGCUUAUUUUGGUCCUUU--UUUAUGCGCC .(((((((.(((((..(((((((((((.((((....)))).-)))))))))))..(..((((...))))..)((((((......))))))((((......)))).)))--)).))))))) ( -49.60) >consensus GGGGACAUUGAAAAUAUUCCGAGCUGCAU______GCAGCUUUUGGAAAAAGGACGGCAUAGUUGCUGUUGCAAUUUCUGUUUUGAAAUUGGCUUAUUUACGUA_UUU__UUUAUGCGCC (....)..........(((((((((((........)))))))..)))).((((((((((....))))))).(((((((......))))))).)))......................... (-23.88 = -22.37 + -1.52)



| Location | 14,753,763 – 14,753,874 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -25.14 |
| Consensus MFE | -12.81 |
| Energy contribution | -14.28 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14753763 111 - 22407834 GGCGCAUAAA--AAA-UACGUAAAUAAGCCAAUUUCAAAACAGAAAUUGCAACGGCAACUAUGCCGUCCUUUUUCCAAAAGCUGA------AUGCAGCCCGGAAUAUUCUCAAUGUCCCC ((.((((...--...-...........((.((((((......)))))))).((((((....)))))).....((((....((((.------...))))..))))........)))).)). ( -25.30) >DroSec_CAF1 17946 110 - 1 UUCGCAUAAA--AAA-UACGUAAAUAAGCCAAUUUCAAAACAGAAAUUGCAACAGCAACUAUGCCGUCCUUUUUCCA-AAGCUGC------AUGCAGCUCGGAAUAUUCUCAAUGUCCCC ...((((...--...-.(((.......((.((((((......))))))))....(((....)))))).....((((.-.(((((.------...))))).))))........)))).... ( -19.60) >DroSim_CAF1 17712 111 - 1 GGCGCAUAAA--AAA-UACGUAAAUAAGCCAAUUUCAAAACAGAAAUUGCAACAGCAACUAUGCCGUCCUUUUUCCAAAAGCUGC------AUGCAGCUCGGAAUAUUCUCAAUGUCCCC ((.((((...--...-.(((.......((.((((((......))))))))....(((....)))))).....((((...(((((.------...))))).))))........)))).)). ( -22.40) >DroEre_CAF1 18531 113 - 1 GGCGCAGAAAAAAAA-UACGUAAAUAAGCCAAUUACAAAACAGAAAUUGCAACAGCAACUAUGCCGCCCUCUUACCAAAAGCCGC------AUGCAACUCGCUAUAUUUUCAAUGUCCCC ((((..(((((....-...((((.........))))......((..((((....)))).(((((.((.............)).))------)))....))......)))))..))))... ( -13.42) >DroYak_CAF1 20066 111 - 1 GGCGCAGAAA--AAA-UACGUAAAUAAGCCAAUUACAAAACAGAAAUUGCAACAGCAACUGUGCCUUUCUCUCCCCAAGAGCUGC------AUGCAGCUCGGAAUGUUUUCAAUGUCCCC (((((((...--...-...((((.........))))..........((((....))))))))))).............((((((.------...))))))(((.((....))...))).. ( -28.50) >DroAna_CAF1 37410 117 - 1 GGCGCAUAAA--AAAGGACCAAAAUAAGCCAAUUUCAAAACAGAAAUUGCAACAGAAACUGCCACAUCCCCCUGCCAC-UGACACAAAUAUCGGUGGCAGGGGACAUUUUCAAUGCACCC ((.((((..(--(((((............(((((((......)))))))............)).....((((((((((-(((........)))))))))))))...))))..)))).)). ( -41.65) >consensus GGCGCAUAAA__AAA_UACGUAAAUAAGCCAAUUUCAAAACAGAAAUUGCAACAGCAACUAUGCCGUCCUCUUUCCAAAAGCUGC______AUGCAGCUCGGAAUAUUCUCAAUGUCCCC ((.((((....................((.((((((......))))))))......................((((...((((((........)))))).))))........)))).)). (-12.81 = -14.28 + 1.47)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:02 2006