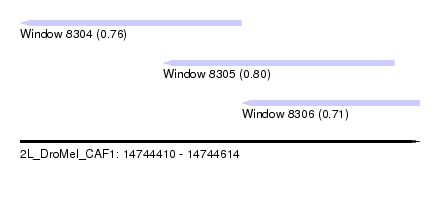
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,744,410 – 14,744,614 |
| Length | 204 |
| Max. P | 0.795811 |

| Location | 14,744,410 – 14,744,523 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.14 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -22.16 |
| Energy contribution | -23.05 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762758 |
| Prediction | RNA |
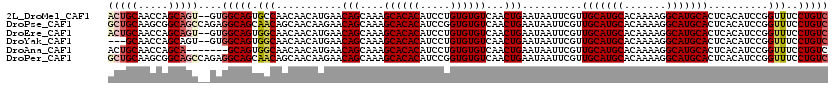
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14744410 113 - 22407834 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUUGCUUUCUGCUUGCCAGAGACAGAAAGGGCUAACCAACACUGUUACACUACAGUGCUGGGAGUGG------ ((((((.......))))))...(((.(((((((....(((...(((((((.((((((.....)))))))))))))))).))))..((((((......))))))...)))))).------ ( -39.60) >DroPse_CAF1 22842 94 - 1 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUU------UGCAUGACAGACACAGAGGCAA--AGGCAAUA----------GCAG---UGGCAGUGG----CA ((((((.......))))))..((((..((.((((((((((((.(.....------.).))))))).....)))))..--.)).....----------((..---..)).))))----.. ( -26.40) >DroSec_CAF1 8588 113 - 1 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUUGCAUUCUGCUUGCCAGAGAUAGAAAGGGCUAACCAACACUGUUACACUGCAGUGCUGGGAGUGG------ ((((((.......))))))...(((.(((((((((((.((..(((((..(((.......))).)))))..)).))))..))))..((((((......))))))...)))))).------ ( -37.80) >DroSim_CAF1 8225 113 - 1 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUUGCUUUCUGCUUGCCAGAGACAGAAAGGGCUAACCAACACUGUUACACUGCAGUGCUGGGAGUGG------ ((((((.......))))))...(((.(((((((....(((...(((((((.((((((.....)))))))))))))))).))))..((((((......))))))...)))))).------ ( -39.90) >DroEre_CAF1 8339 119 - 1 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUUGCUUUCUGCUUGCCAGAGACAGAAAGGGCUAACCAACACUGUUGCAGUGCAGUGCUGGCAGUGGAGGGGA ((((((.......))))))..............((((.((..((((((((.((((((.....))))))))))))))(((..(((.((((((......)))))).))).))).)).)))) ( -41.40) >DroPer_CAF1 22692 94 - 1 GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUU------UGCAUGACAGACACAGAGGCAA--AGGCAAUA----------GCAG---UGGCAGUGG----CA ((((((.......))))))..((((..((.((((((((((((.(.....------.).))))))).....)))))..--.)).....----------((..---..)).))))----.. ( -26.40) >consensus GCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUCAUUCUUUUUGCUUUCUGCUUGCCAGAGACAGAAAGGGCUAACCAACACUGUUACACUGCAGUGCUGGCAGUGG______ ((((((.......))))))...(((..(((((...((((..(((((((((...((((.....))))..)))))))))....................)))).)))))..)))....... (-22.16 = -23.05 + 0.89)

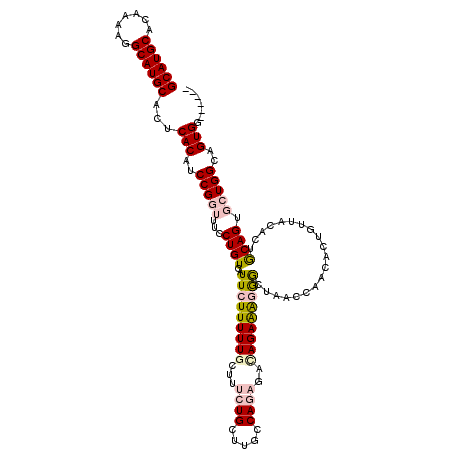
| Location | 14,744,483 – 14,744,601 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -28.13 |
| Energy contribution | -28.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14744483 118 - 22407834 ACUGCAACCAGCAGU--GUGGCAGUGCCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((.....)))))--..(((((.(((...........(((....((((((.....))))))...)))..........(((((((.......))))))).........)))...))))) ( -34.20) >DroPse_CAF1 22896 120 - 1 GCUGCAAGCGGCAGCCAGAGGCAGCAACAGCAACAAGAACAGCAAAGCACACAUCCGGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((..((....)).....))))).((((.(((..((.(((....((((((.....))))))...)))..........(((((((.......)))))))......))..)))..)))). ( -34.20) >DroEre_CAF1 8418 118 - 1 ACUGCAACCAGCAGU--GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((.....)))))--..(((((.((.(((...(((..(((....((((((.....))))))...)))..........(((((((.......)))))))....)))...)))))))))) ( -33.90) >DroYak_CAF1 9962 115 - 1 ---GCAACCAGCAGU--GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC ---..((((.(..((--((((((.((....))...))..(((....((((((.....))))))...)))..........(((((((.......))))))).))))))).))))....... ( -29.50) >DroAna_CAF1 29233 113 - 1 ACUGCAACCAGCA-------GCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((........-------)))))((.(((...(((..(((....((((((.....))))))...)))..........(((((((.......)))))))....)))...))).)).... ( -30.60) >DroPer_CAF1 22746 120 - 1 GCUGCAAGCGGCAGCCAGAGGCAGCAACAGCAACAAGAACAGCAAAGCACACAUCCGGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((..((....)).....))))).((((.(((..((.(((....((((((.....))))))...)))..........(((((((.......)))))))......))..)))..)))). ( -34.20) >consensus ACUGCAACCAGCAGU__GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUUGCAUGCACAAAAGGCAUGCACUCACAUCCGGUUUCCUGUC (((((.....)))))....(((((.(((...........(((....((((((.....))))))...)))..........(((((((.......)))))))..........)))..))))) (-28.13 = -28.68 + 0.56)

| Location | 14,744,523 – 14,744,614 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -14.05 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14744523 91 - 22407834 --------------CCAGAAGCCA-------GCAACUGCAACCAGCAGU--GUGGCAGUGCCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUU --------------...(((((((-------...(((((.....)))))--.)))).................(((....((((((.....))))))...))).....)))... ( -24.30) >DroPse_CAF1 22936 92 - 1 ---------------CACAAGCGG-------ACUGCUGCAAGCGGCAGCCAGAGGCAGCAACAGCAACAAGAACAGCAAAGCACACAUCCGGUGUGUCAACUGAAUAAUUCGUU ---------------....(((((-------.((((((....)))))))).(((...((....))........(((....((((((.....))))))...))).....)))))) ( -24.50) >DroEre_CAF1 8458 98 - 1 --------------CCAGCAGCCAACAGCCAGCAACUGCAACCAGCAGU--GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUU --------------..(((.((((...((((...(((((.....)))))--.))))..))))...........(((....((((((.....))))))...)))........))) ( -30.30) >DroYak_CAF1 10002 85 - 1 --------------CCAGCAGCCA-------------GCAACCAGCAGU--GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUU --------------..(((.((((-------------.(..(((.....--.)))..)))))...........(((....((((((.....))))))...)))........))) ( -18.20) >DroAna_CAF1 29273 107 - 1 GCGGCAACUGGCAACCAGCAACCGGCAACCAGCAACUGCAACCAGCA-------GCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUU .(((...((((...))))...))).......((.(((((........-------))))).))...........(((....((((((.....))))))...)))........... ( -26.60) >DroPer_CAF1 22786 92 - 1 ---------------CACAAGCGG-------ACUGCUGCAAGCGGCAGCCAGAGGCAGCAACAGCAACAAGAACAGCAAAGCACACAUCCGGUGUGUCAACUGAAUAAUUCGUU ---------------....(((((-------.((((((....)))))))).(((...((....))........(((....((((((.....))))))...))).....)))))) ( -24.50) >consensus ______________CCAGAAGCCA_______GCAACUGCAACCAGCAGU__GUGGCAGUGGCAACAACAUGAACAGCAAAGCACACAUCCUGUGUGUCAACUGAAUAAUUCGUU ..................................(((((.....)))))........((....))........(((....((((((.....))))))...)))........... (-14.05 = -14.22 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:54 2006