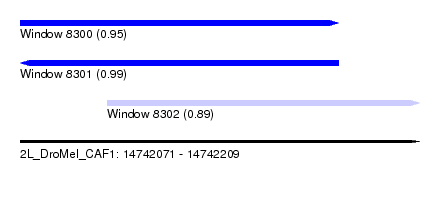
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,742,071 – 14,742,209 |
| Length | 138 |
| Max. P | 0.991745 |

| Location | 14,742,071 – 14,742,181 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
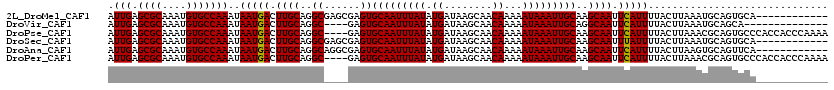
>2L_DroMel_CAF1 14742071 110 + 22407834 -------UGGAGGA--CGU-UGUAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCGAGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA -------....(.(--(((-(((....))))))).).(((((((.((((....))))....)))))))((((......)))).((((((((((.((........))...)))))))))). ( -29.00) >DroVir_CAF1 14883 91 + 1 -------------------------GAGCAAUGUGCAGCCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAG -------------------------..((((.((.((....(((.((((....)))))))....))))))))....----...((((((((((.((........))...)))))))))). ( -20.70) >DroPse_CAF1 20018 108 + 1 -------UGAAGGAGCAGC-AGCAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA -------.......((.((-((((.(.....).)))((((((((.((((....))))....)))))))))))..))----...((((((((((.((........))...)))))))))). ( -26.60) >DroEre_CAF1 6487 110 + 1 -------UGGAGGA--CGA-UGUAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCGAGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA -------....(.(--((.-(((....))).))).).(((((((.((((....))))....)))))))((((......)))).((((((((((.((........))...)))))))))). ( -25.00) >DroAna_CAF1 27564 118 + 1 AGUGGCCGGGAAGA--UGUAAGUAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCAGGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA .((.(((.......--(((........)))...(((((((((((.((((....))))....))))))).)))))))..))...((((((((((.((........))...)))))))))). ( -30.00) >DroPer_CAF1 19860 108 + 1 -------UGAAGGAGCAGC-AGCAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA -------.......((.((-((((.(.....).)))((((((((.((((....))))....)))))))))))..))----...((((((((((.((........))...)))))))))). ( -26.60) >consensus _______UGGAGGA__AGC_AGUAGAAACAAUGUGCAGUCAUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC____GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAA ...............................(.(((((((((((.((((....))))....))))))).)))).)........((((((((((.((........))...)))))))))). (-22.67 = -22.83 + 0.17)

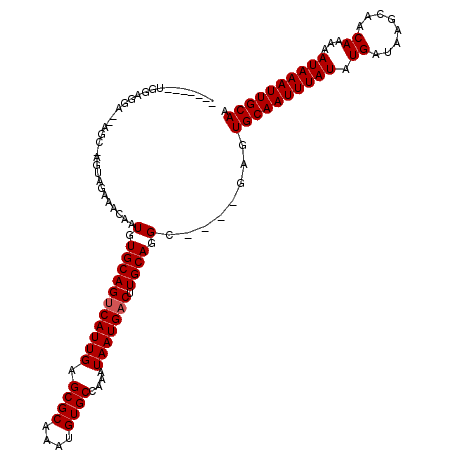
| Location | 14,742,071 – 14,742,181 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -22.98 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14742071 110 - 22407834 UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUCGCUCGCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUACA-ACG--UCCUCCA------- .((((((((((...((........)).))))))))))...........(((.((((((....(((.(....).))).)))))))))((.((((....)))-).)--)......------- ( -22.30) >DroVir_CAF1 14883 91 - 1 CUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUC----GCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGGCUGCACAUUGCUC------------------------- .((((((((((...((........)).))))))))))...----((.((((.((((((....(((.(....).))).))))))))))....))..------------------------- ( -21.50) >DroPse_CAF1 20018 108 - 1 UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUC----GCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUGCU-GCUGCUCCUUCA------- .((((((((((...((........)).))))))))))...----((..((((((((((....(((.(....).))).)))))))(((.........))))-)).)).......------- ( -23.40) >DroEre_CAF1 6487 110 - 1 UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUCGCUCGCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUACA-UCG--UCCUCCA------- .((((((((((...((........)).))))))))))..........((((.((((((....(((.(....).))).)))))))))).............-...--.......------- ( -21.40) >DroAna_CAF1 27564 118 - 1 UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUCGCCUGCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUACUUACA--UCUUCCCGGCCACU .((((((((((...((........)).))))))))))...(((....((((.((((((....(((.(....).))).))))))))))...(((........)))--.......))).... ( -25.90) >DroPer_CAF1 19860 108 - 1 UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUC----GCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUGCU-GCUGCUCCUUCA------- .((((((((((...((........)).))))))))))...----((..((((((((((....(((.(....).))).)))))))(((.........))))-)).)).......------- ( -23.40) >consensus UUGCAAUUUAUUUUUGUUGCUUAUCAUAUAAAUUGCACUC____GCCUGCAAGUCAUUAUUUGGCACAUUUGCGCUCAAUGACUGCACAUUGUUUCUACU_ACG__UCCUCCA_______ .((((((((((...((........)).))))))))))..........((((.((((((....(((.(....).))).))))))))))................................. (-21.42 = -21.28 + -0.14)



| Location | 14,742,101 – 14,742,209 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.99 |
| Mean single sequence MFE | -22.89 |
| Consensus MFE | -20.22 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14742101 108 + 22407834 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCGAGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUCAUUUUACUUAAAUGCAGUGCA------------ .(((.((((....)))))))......((((((......))))))(((((((((.((........))...)))))))))..(((...(((((.....)))))...))).------------ ( -26.60) >DroVir_CAF1 14898 102 + 1 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAGGCAAUUCAUUUUACUUAAAUGCAGCA-------------- .(((((...(((((((((........(((((....)----))))(((((((((.((........))...))))))))).))))...)))))..)))))........-------------- ( -21.60) >DroPse_CAF1 20050 116 + 1 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUCAUUUUACUUAAACGCAGUGCCCACCACCCAAAA ..((.((((...((((......(((((.((((....----...((((((((((.((........))...)))))))))).)))).))))).........)))))))).)).......... ( -20.76) >DroSec_CAF1 6628 108 + 1 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCGAGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUUAUUUUACUUAAAUGCAGUGCA------------ .....((((...(((((((.((((((((((((......)))))).))..)))).))....)).)))(((((((((((....)))))))))))...........)))).------------ ( -24.90) >DroAna_CAF1 27602 108 + 1 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGCAGGCGAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUCAUUUUACUUAAGUGCAGUUCA------------ .(((.((((....)))))))......((((((......))))))(((((((((.((........))...)))))))))..(.(((((((((.....))))).))))).------------ ( -22.70) >DroPer_CAF1 19892 116 + 1 AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC----GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUCAUUUUACUUAAACGCAGUGCCCACCACCCAAAA ..((.((((...((((......(((((.((((....----...((((((((((.((........))...)))))))))).)))).))))).........)))))))).)).......... ( -20.76) >consensus AUUGAGCGCAAAUGUGCCAAAUAAUGACUUGCAGGC____GAGUGCAAUUUAUAUGAUAAGCAACAAAAAUAAAUUGCAAGCAAUUCAUUUUACUUAAAUGCAGUGCA____________ .(((.((((....)))))))..(((((.((((..((......))(((((((((.((........))...)))))))))..)))).))))).............................. (-20.22 = -20.08 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:50 2006