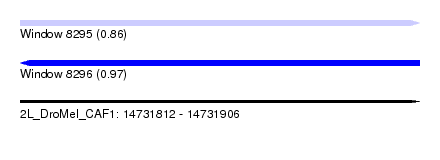
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,731,812 – 14,731,906 |
| Length | 94 |
| Max. P | 0.972186 |

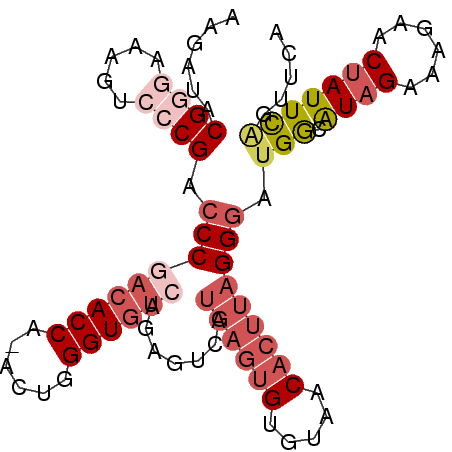
| Location | 14,731,812 – 14,731,906 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -26.88 |
| Consensus MFE | -14.38 |
| Energy contribution | -17.02 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14731812 94 + 22407834 UGAACGGAAUAGUUCUUUCUAUGCCAUCCCUAAGUGUUACACACUCAUGACUCUGACACCCAGU-UGGUGUCGGGUCGGGACUUUCCCGUAUCUU .((((((.((((......))))....((((..((((.....))))...(((.((((((((....-.))))))))))))))).....)))).)).. ( -32.00) >DroSec_CAF1 88665 95 + 1 UGGACUGAAUAGUUCUUUCUACGCCAUUCCUAAGUGUUACACACUCAUGACUCCGACACCCAGUUUGGUGUCGGGUCGGGACUUUCCCGUGUCUU .(((.((..(((......)))...)).)))........((((......(((.((((((((......)))))))))))(((.....)))))))... ( -31.70) >DroSim_CAF1 89362 95 + 1 UGAACUGAAUAGUUCUUUCUAUGCCAUCCCUAAGUGUUACACACUCAUGACUCCGACACCCAGUUUGGUGUCGGGUCGGGACUUUCCCGUAUCUU .(((((....)))))...........((((..((((.....))))...(((.((((((((......))))))))))))))).............. ( -31.30) >DroEre_CAF1 91801 73 + 1 ----CCAAAUAGUGUCCUCGAUGCCAUCCCUAAGUGUUACAC---------------ACCCAGU-UGGUGUCGGGUCG--ACUUUCUCGUAUCUU ----.........(((((((((((((...((..(((.....)---------------))..)).-))))))))))..)--))............. ( -18.50) >DroYak_CAF1 88479 91 + 1 UGAACUGAAUAGUUU-UUCUAUGUCAUCCCUAAGUGUUACACACUCAUGACUCUGACACCCAGU-UGGUGUGGGGUCG--ACUUUCCCGUAUCUU .(((((....)))))-......(((.......((((.....))))...((((((.(((((....-.))))))))))))--))............. ( -20.90) >consensus UGAACUGAAUAGUUCUUUCUAUGCCAUCCCUAAGUGUUACACACUCAUGACUCCGACACCCAGU_UGGUGUCGGGUCGGGACUUUCCCGUAUCUU .((((......))))................(((...(((........(((.((((((((......)))))))))))(((.....)))))).))) (-14.38 = -17.02 + 2.64)

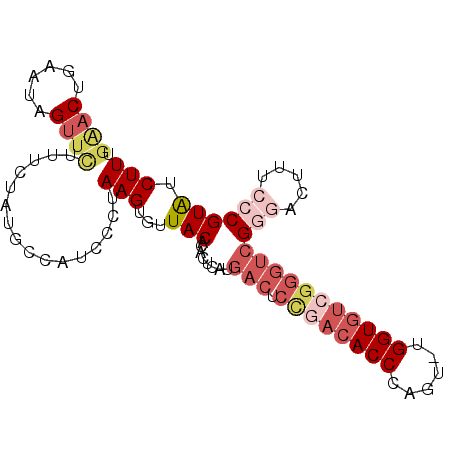
| Location | 14,731,812 – 14,731,906 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.96 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -16.18 |
| Energy contribution | -18.78 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14731812 94 - 22407834 AAGAUACGGGAAAGUCCCGACCCGACACCA-ACUGGGUGUCAGAGUCAUGAGUGUGUAACACUUAGGGAUGGCAUAGAAAGAACUAUUCCGUUCA ..((.((((((..(((((((((.((((((.-....)))))).).))).((((((.....)))))))))))....(((......))))))))))). ( -35.10) >DroSec_CAF1 88665 95 - 1 AAGACACGGGAAAGUCCCGACCCGACACCAAACUGGGUGUCGGAGUCAUGAGUGUGUAACACUUAGGAAUGGCGUAGAAAGAACUAUUCAGUCCA ..(((.((((.....))))..((((((((......)))))))).))).((((((.....))))))(((.(((.((((......))))))).))). ( -38.50) >DroSim_CAF1 89362 95 - 1 AAGAUACGGGAAAGUCCCGACCCGACACCAAACUGGGUGUCGGAGUCAUGAGUGUGUAACACUUAGGGAUGGCAUAGAAAGAACUAUUCAGUUCA .........(...((((((((((((((((......)))))))).))).((((((.....)))))))))))..).......(((((....))))). ( -38.40) >DroEre_CAF1 91801 73 - 1 AAGAUACGAGAAAGU--CGACCCGACACCA-ACUGGGU---------------GUGUAACACUUAGGGAUGGCAUCGAGGACACUAUUUGG---- ......(.(((.(((--.(.(((((..(((-.((((((---------------(.....)))))))...)))..))).)).)))).))).)---- ( -18.90) >DroYak_CAF1 88479 91 - 1 AAGAUACGGGAAAGU--CGACCCCACACCA-ACUGGGUGUCAGAGUCAUGAGUGUGUAACACUUAGGGAUGACAUAGAA-AAACUAUUCAGUUCA ..((.((..(((.((--((.(((.(((((.-....)))))........((((((.....))))))))).)))).(((..-...)))))).)))). ( -22.70) >consensus AAGAUACGGGAAAGUCCCGACCCGACACCA_ACUGGGUGUCAGAGUCAUGAGUGUGUAACACUUAGGGAUGGCAUAGAAAGAACUAUUCAGUUCA ......((((.....)))).(((((((((......)))))).......((((((.....))))))))).(((.((((......)))))))..... (-16.18 = -18.78 + 2.60)

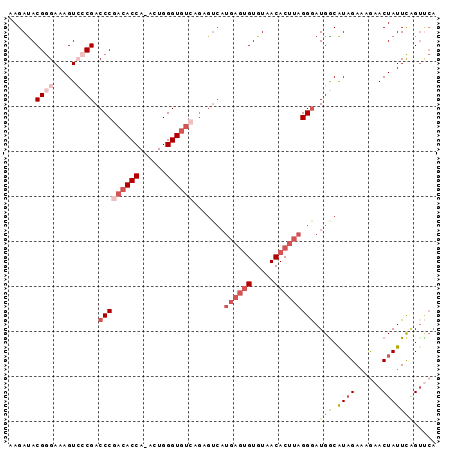
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:45 2006