
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,588,167 – 1,588,263 |
| Length | 96 |
| Max. P | 0.669463 |

| Location | 1,588,167 – 1,588,263 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -29.38 |
| Consensus MFE | -22.45 |
| Energy contribution | -21.59 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
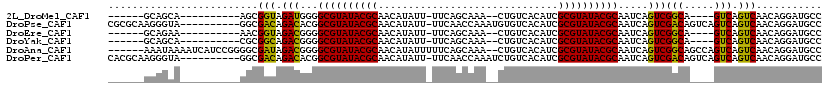
>2L_DroMel_CAF1 1588167 96 + 22407834 ------GCAGCA----------AGCGGUAGAUGGGGCGUAUACGCAACAUAUU-UUCAGCAAA--CUGUCACAUCGCGUAUACGCAAUCAGUCGGCA----GUCAGUCAACAGGAUGCC ------((....----------.))((..(((...((((((((((........-..(((....--))).......)))))))))).)))..))((((----..(........)..)))) ( -26.83) >DroPse_CAF1 110506 108 + 1 CGCGCAAGGGUA----------GGCGACAGACACGGCGUAUACGCAACAUAUU-UUCAACCAAAUGUGUCACAUCGCGUAUACGCAAUCAGUCGACAGUCAGUCAGUCAACAGGAUGCC ...(((...((.----------((((((.((....((((((((((.(((((((-(......))))))))......))))))))))..)).)))(((.....))).))).))....))). ( -33.10) >DroEre_CAF1 68169 96 + 1 ------GCAGAA----------AACGGUAGACGGGGCGUAUACGCAACAUAUU-UUCAGCAAA--CUGUCACAUCGCGUAUACGCAAUCAGUCGGCA----GUCAGUCAACAGGAUGCC ------......----------.......(((...((((((((((........-..(((....--))).......)))))))))).....)))((((----..(........)..)))) ( -26.03) >DroYak_CAF1 66829 96 + 1 ------GCAGCA----------CGCGGCAGACGGGGCGUAUACGCAACAUAUU-UUCAGCAAA--CUGUCACAUCGCGUAUACGCAAUCAGUCGGCA----GUCAGUCAACAGGAUGCC ------((....----------.))(((((((...((((((((((........-..(((....--))).......)))))))))).....)))(((.----....))).......)))) ( -28.93) >DroAna_CAF1 97476 111 + 1 ------AAAUAAAAUCAUCCGGGGCGAUAGACGGGGCGUAUACGCAACAUAUUUUUCAGCAAA--CUGUCACAUCGCGUAUACGCAAUCAGUCGGCAGCCAGUCAGUCAACAGGAUGCC ------.........(((((..(((....(((...((((((((((...........(((....--))).......)))))))))).....)))....))).((......)).))))).. ( -32.37) >DroPer_CAF1 113355 108 + 1 CACGCAAGGGUA----------GGCGACAGACACGGCGUAUACGCAACAUAUU-UUCAACCAAAUCUGUCACAUCGCGUAUACGCAAUCAGUCGACAGUCAGUCAGUCAACAGGAUGCC ...(((...((.----------((((((.((....((((((((((.(((.(((-(......)))).)))......))))))))))..)).)))(((.....))).))).))....))). ( -29.00) >consensus ______ACAGAA__________GGCGACAGACGGGGCGUAUACGCAACAUAUU_UUCAGCAAA__CUGUCACAUCGCGUAUACGCAAUCAGUCGGCA____GUCAGUCAACAGGAUGCC .........................(((.(((...((((((((((..............................)))))))))).....)))(((.....))).)))........... (-22.45 = -21.59 + -0.86)
| Location | 1,588,167 – 1,588,263 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.23 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
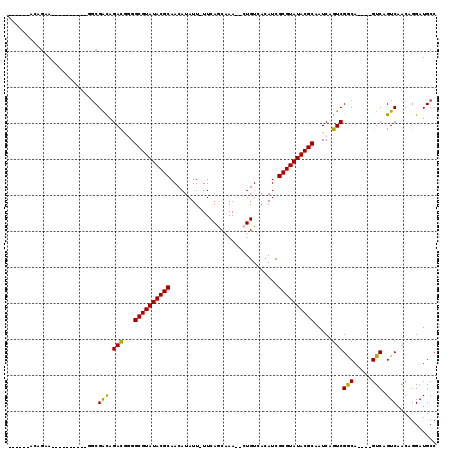
>2L_DroMel_CAF1 1588167 96 - 22407834 GGCAUCCUGUUGACUGAC----UGCCGACUGAUUGCGUAUACGCGAUGUGACAG--UUUGCUGAA-AAUAUGUUGCGUAUACGCCCCAUCUACCGCU----------UGCUGC------ ((((..(.((((.(....----.).)))).(((.((((((((((((((((.(((--....)))..-..))))))))))))))))...)))....)..----------))))..------ ( -30.00) >DroPse_CAF1 110506 108 - 1 GGCAUCCUGUUGACUGACUGACUGUCGACUGAUUGCGUAUACGCGAUGUGACACAUUUGGUUGAA-AAUAUGUUGCGUAUACGCCGUGUCUGUCGCC----------UACCCUUGCGCG (((............(((.....)))(((.(((.((((((((((((((((...((......))..-..))))))))))))))))...))).))))))----------............ ( -35.60) >DroEre_CAF1 68169 96 - 1 GGCAUCCUGUUGACUGAC----UGCCGACUGAUUGCGUAUACGCGAUGUGACAG--UUUGCUGAA-AAUAUGUUGCGUAUACGCCCCGUCUACCGUU----------UUCUGC------ ((((..(........)..----))))(((.(...((((((((((((((((.(((--....)))..-..)))))))))))))))).).))).......----------......------ ( -31.10) >DroYak_CAF1 66829 96 - 1 GGCAUCCUGUUGACUGAC----UGCCGACUGAUUGCGUAUACGCGAUGUGACAG--UUUGCUGAA-AAUAUGUUGCGUAUACGCCCCGUCUGCCGCG----------UGCUGC------ (((((.(.(((....)))----.((.(((.(...((((((((((((((((.(((--....)))..-..)))))))))))))))).).))).)).).)----------))))..------ ( -32.80) >DroAna_CAF1 97476 111 - 1 GGCAUCCUGUUGACUGACUGGCUGCCGACUGAUUGCGUAUACGCGAUGUGACAG--UUUGCUGAAAAAUAUGUUGCGUAUACGCCCCGUCUAUCGCCCCGGAUGAUUUUAUUU------ ((((.((.(((....))).)).))))........((((((((((((((((.(((--....))).....))))))))))))))))..(((((........))))).........------ ( -37.10) >DroPer_CAF1 113355 108 - 1 GGCAUCCUGUUGACUGACUGACUGUCGACUGAUUGCGUAUACGCGAUGUGACAGAUUUGGUUGAA-AAUAUGUUGCGUAUACGCCGUGUCUGUCGCC----------UACCCUUGCGUG (((............(((.....)))(((.(((.((((((((((((((((.(((......)))..-..))))))))))))))))...))).))))))----------............ ( -34.80) >consensus GGCAUCCUGUUGACUGAC____UGCCGACUGAUUGCGUAUACGCGAUGUGACAG__UUUGCUGAA_AAUAUGUUGCGUAUACGCCCCGUCUACCGCC__________UACUGC______ ((((....(((....)))....))))(((.....((((((((((((((((.(((......))).....))))))))))))))))...)))............................. (-27.86 = -27.87 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:15 2006