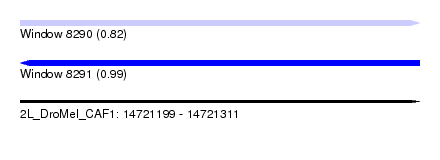
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,721,199 – 14,721,311 |
| Length | 112 |
| Max. P | 0.991821 |

| Location | 14,721,199 – 14,721,311 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14721199 112 + 22407834 UAUAAAAAGUUUUUCCCUUCUACGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUGUGCUGCUUAUGCCACAGUGCUUCCUUCGCUUUUCUUUGCU .(((((((.....(((.......)))....)))))))((...(((....((((((.(((.......))).))))))((....))..))).)).......((........)). ( -19.30) >DroSec_CAF1 77998 110 + 1 UAUAAAAAGUU-UUCCCUUCUACGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUGUGCUGCUUAUGCCACAGUGCUUCCUUUGCUUCU-UCUGCU ((((((((...-.(((.......)))....))))))))...........((((...((........((((((((..((....))))))).)))......))....-..)))) ( -19.94) >DroSim_CAF1 78117 111 + 1 UAUAAAAAGUUUUUCCCUUCUACGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUGUGCUGCUUAUGCCACAGUGCUUCCUUCGCUUCU-UCUGCU .(((((((.....(((.......)))....)))))))((...(((....((((((.(((.......))).))))))((....))..))).)).......((....-...)). ( -19.00) >DroEre_CAF1 81050 110 + 1 UAUAAAAAGUU-UUCCCUUCUUCGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUGUGCUGCUUAUGCCACCGAGCAUCCUUCGCCUUU-UCCGCU ((((((((...-.(((.......)))....))))))))......((...((((((.(((.......))).))))))((((........)))).............-...)). ( -19.80) >DroYak_CAF1 77190 110 + 1 UAUAAAAAGUU-UUCCCUUCUUCGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUCUGCUGCUUAUGCCACCGUGCAUCCUUCGCCUCU-UCUGUU ((((((((...-.(((.......)))....))))))))......((..(((((...(((.......)))...)))))...((((......)))).....))....-...... ( -14.80) >consensus UAUAAAAAGUU_UUCCCUUCUACGGAUUUAUUUUUAUGCCUUCUGCCUUAGCAUAUGCUUGUUUAUAGCCUGUGCUGCUUAUGCCACAGUGCUUCCUUCGCUUCU_UCUGCU ((((((((.....(((.......)))....))))))))......(((..((((((.(((.......))).))))))((....))....).)).......((........)). (-16.40 = -16.44 + 0.04)


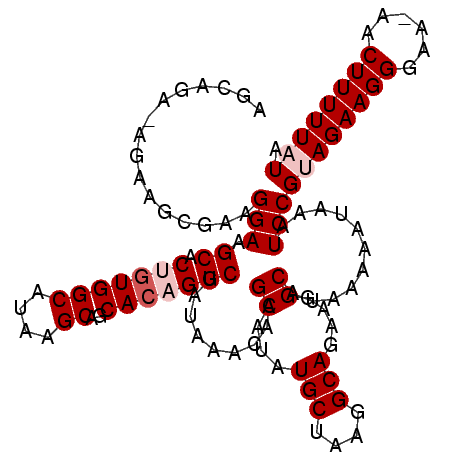
| Location | 14,721,199 – 14,721,311 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -20.04 |
| Energy contribution | -21.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14721199 112 - 22407834 AGCAAAGAAAAGCGAAGGAAGCACUGUGGCAUAAGCAGCACAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGUAGAAGGGAAAAACUUUUUAUA .((........))...((((((.(((((((....))..))))))))........((...(((....)))....))...........)))((((((((......)))))))). ( -26.80) >DroSec_CAF1 77998 110 - 1 AGCAGA-AGAAGCAAAGGAAGCACUGUGGCAUAAGCAGCACAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGUAGAAGGGAA-AACUUUUUAUA .((...-....))...((((((.(((((((....))..))))))))........((...(((....)))....))...........)))((((((((...-..)))))))). ( -26.30) >DroSim_CAF1 78117 111 - 1 AGCAGA-AGAAGCGAAGGAAGCACUGUGGCAUAAGCAGCACAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGUAGAAGGGAAAAACUUUUUAUA .((...-....))...((((((.(((((((....))..))))))))........((...(((....)))....))...........)))((((((((......)))))))). ( -26.50) >DroEre_CAF1 81050 110 - 1 AGCGGA-AAAGGCGAAGGAUGCUCGGUGGCAUAAGCAGCACAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGAAGAAGGGAA-AACUUUUUAUA ..((((-.....(....).((((...(((((((.(((((....)))........)).))))))).)))).................))))((((((....-..))))))... ( -23.20) >DroYak_CAF1 77190 110 - 1 AACAGA-AGAGGCGAAGGAUGCACGGUGGCAUAAGCAGCAGAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGAAGAAGGGAA-AACUUUUUAUA ....((-((((((......(((....(((((((.(((((....)))........)).)))))))..)))....))...........(((......)))..-..))))))... ( -21.50) >consensus AGCAGA_AGAAGCGAAGGAAGCACUGUGGCAUAAGCAGCACAGGCUAUAAACAAGCAUAUGCUAAGGCAGAAGGCAUAAAAAUAAAUCCGUAGAAGGGAA_AACUUUUUAUA ................(((.((.(((((((....))..))))))).........((...(((....)))....))...........)))((((((((......)))))))). (-20.04 = -21.04 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:40 2006