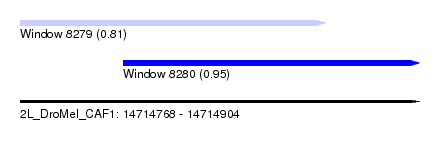
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,714,768 – 14,714,904 |
| Length | 136 |
| Max. P | 0.946180 |

| Location | 14,714,768 – 14,714,872 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 97.38 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
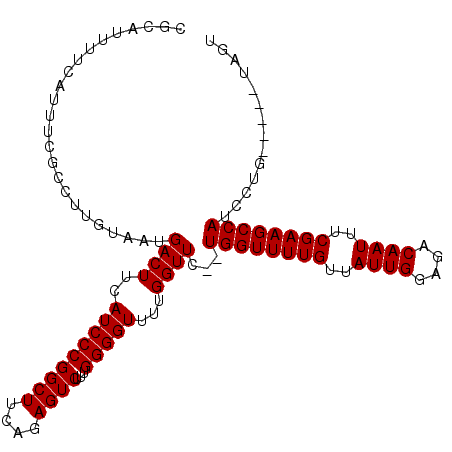
>2L_DroMel_CAF1 14714768 104 + 22407834 CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGU .(((....................((((..(((((((((....))))...)))))...)))).--((((((((..((((....))))..))))))))...))-----)... ( -26.30) >DroSec_CAF1 71824 104 + 1 CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGU .(((....................((((..(((((((((....))))...)))))...)))).--((((((((..((((....))))..))))))))...))-----)... ( -26.30) >DroSim_CAF1 71774 104 + 1 CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGU .(((....................((((..(((((((((....))))...)))))...)))).--((((((((..((((....))))..))))))))...))-----)... ( -26.30) >DroEre_CAF1 71379 106 + 1 CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUCUUUGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGU .(((....................((((..(((((((((....))))...)))))...))))...((((((((..((((....))))..))))))))...))-----)... ( -26.40) >DroYak_CAF1 70871 109 + 1 CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUGUCCCGUAGU ..............(((..(..(((....)))..))))........((.(((((....((...--((((((((..((((....))))..)))))))).))..))))).)). ( -28.60) >consensus CGCAUUUUCAUUUCGCCUUGUAAUGACUUCAUCCCGGCUUCAGAGUCUUUGGGGUUUUGGUUC__UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG_____UAGU ........................((((..(((((((((....))))...)))))...))))...((((((((..((((....))))..)))))))).............. (-25.54 = -25.54 + -0.00)

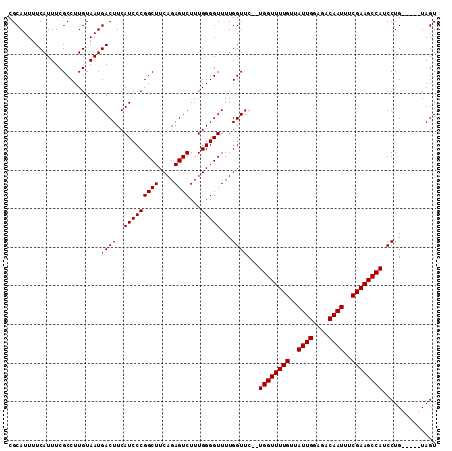
| Location | 14,714,803 – 14,714,904 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
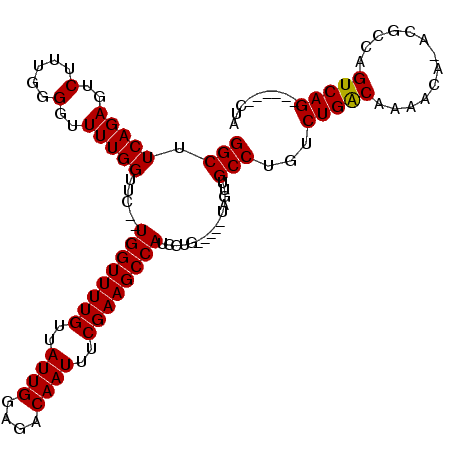
>2L_DroMel_CAF1 14714803 101 + 22407834 GGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGUUGCCUGUCUGACAAAACA-ACGCCAGUCAG------CUA (((.(((((..(.....)..)))))...--((((((((..((((....))))..)))))))).....-----.....))).(.(((((......-......)))))------).. ( -28.90) >DroSec_CAF1 71859 101 + 1 GGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGUUGCCUGUCUGACAAAACA-ACGCCAGUCAG------CUA (((.(((((..(.....)..)))))...--((((((((..((((....))))..)))))))).....-----.....))).(.(((((......-......)))))------).. ( -28.90) >DroSim_CAF1 71809 101 + 1 GGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGUUGCCUGUCUGACAAAACA-ACGCCAGUCAG------CUA (((.(((((..(.....)..)))))...--((((((((..((((....))))..)))))))).....-----.....))).(.(((((......-......)))))------).. ( -28.90) >DroEre_CAF1 71414 104 + 1 GGCUUCAGAGUCUUUGGGGUUUUGGUUCUUUGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG-----UAGUUGCCUGUCUGGCAAAACAAACGCCAGUCAG------CUA (((.(((((..(.....)..))))).....((((((((..((((....))))..)))))))).....-----.....)))((.(((((.........))))).)).------... ( -33.80) >DroYak_CAF1 70906 112 + 1 GGCUUCAGAGUCUUUGGGGUUUUGGUUC--UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUGUCCCGUAGUUGCCAGUCUGACAAAACA-ACGCCAGUCAGCGUUGGCUA (((.(((((.(((.(((((....((...--((((((((..((((....))))..)))))))).))..))))).)).....).))))).....((-((((......))))))))). ( -39.00) >consensus GGCUUCAGAGUCUUUGGGGUUUUGGUUC__UGGUUUUGUUAUUGGAGACAAUUUCGAAGCCAUCCUG_____UAGUUGCCUGUCUGACAAAACA_ACGCCAGUCAG______CUA (((.(((((..(.....)..))))).....((((((((..((((....))))..))))))))...............)))...(((((.............)))))......... (-28.28 = -28.12 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:30 2006