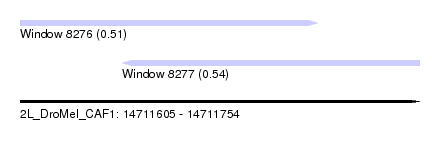
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,711,605 – 14,711,754 |
| Length | 149 |
| Max. P | 0.542426 |

| Location | 14,711,605 – 14,711,716 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -20.84 |
| Consensus MFE | -17.78 |
| Energy contribution | -17.58 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14711605 111 + 22407834 GUGCCCGACUAAACACAAAUUCGAUGAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGCCAAAUAAAUUCAUUUAGGGUUUUAUGCGCUCCACUGAAGUAUGCCAAAAGGCA .(((((((............)))...................(((((((((...((((((...((((((.....))))))..)).))))...))))))).)).....)))) ( -21.00) >DroSec_CAF1 68360 110 + 1 -UGCCCGACUAAACACAAAUUCGAUGAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGUCAAAUAAAUUCAUUUAGGGCUUUAUGCGCUCCACUGAAGUAUGCCAAAAGGCA -.(((........................................(((((((((((((.......)))))))))))))..(((((..((....)).)))))......))). ( -19.60) >DroSim_CAF1 68588 110 + 1 -UGCCCGACUAAACACAAAUUCGAUGAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGCUAAAUAAAUUCAUUUAGGGCUUUAUGCGCUCCACUGAAGUAUGCCAAAAGGCA -.((((((............)))...................(((((((((...((((...(((((..(......)..)))))..))))...))))))).)).....))). ( -19.40) >DroEre_CAF1 67976 110 + 1 -UGCGCGACUAAACACAAAUUCGAUGAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGCCAAAUAAAUUCAUUUAGGGCUUUAUGUGCUCCACUGAAGUGCGCCAAAAGGCA -.((((.......................................((((((((((((.........))))))))))))((((.(((...)))))))))))(((....))). ( -22.20) >DroYak_CAF1 67669 109 + 1 -UGCCCGACUAAACACAAAUUCGAUAAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGCCAAAUAAAUUCAUUUAGGGCUUUAUGCGCUCCACUGAAGUGCGCCAA-AGGCA -.((((((............)))...................((.((((((((((((.........)))))))))))).....(((((.......)))))))...-.))). ( -22.00) >consensus _UGCCCGACUAAACACAAAUUCGAUGAAAUAAAAAUAAAUUAGCUGCUUUAAAUGAGCCAAAUAAAUUCAUUUAGGGCUUUAUGCGCUCCACUGAAGUAUGCCAAAAGGCA ..((((((............)))...................(((((((((...((((...(((((..(......)..)))))..))))...))))))).)).....))). (-17.78 = -17.58 + -0.20)

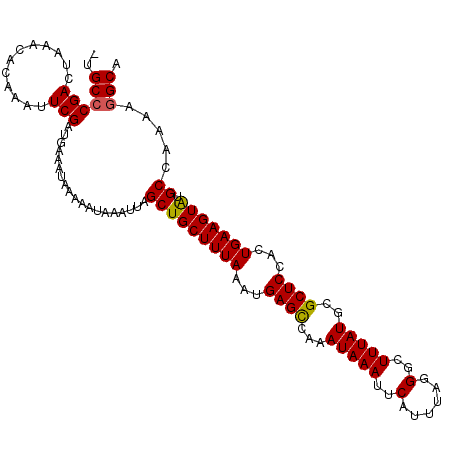
| Location | 14,711,643 – 14,711,754 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -18.38 |
| Energy contribution | -20.38 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14711643 111 - 22407834 A--AAGAGGGUGAUGCGAAAGACUGCGACGACGAAGACGGUGCCUUUUGGCAUACUUC--AGUGGAGCGCAUAAAACCCUAAAUGAAUUUAUUUGGCUCAUUUAA-AGCAGCUAAU .--...(((((.(((((.....).((..((..((((...(((((....))))).))))--..))..))))))...)))))............(((((((......-.).)))))). ( -31.40) >DroSec_CAF1 68397 112 - 1 A-AAAGAGGGUGAUGCGAAUGACUGCGACGACGAAGACGGUGCCUUUUGGCAUACUUC--AGUGGAGCGCAUAAAGCCCUAAAUGAAUUUAUUUGACUCAUUUAA-AGCAGCUAAU .-....(((((.(((((..(.((((((.(......).))(((((....)))))....)--))).)..)))))...)))))((((((.((.....)).))))))..-.......... ( -30.60) >DroSim_CAF1 68625 112 - 1 A-AAAGAGGGUGAUGCGAAAGACUGCGACGACGAAGACGGUGCCUUUUGGCAUACUUC--AGUGGAGCGCAUAAAGCCCUAAAUGAAUUUAUUUAGCUCAUUUAA-AGCAGCUAAU .-....(((((.(((((.....).((..((..((((...(((((....))))).))))--..))..))))))...)))))............(((((((......-.).)))))). ( -32.00) >DroEre_CAF1 68013 113 - 1 AAAAAGAGGGUGUUGCGAAAGCCAGCGAAGACGAAGACGGUGCCUUUUGGCGCACUUC--AGUGGAGCACAUAAAGCCCUAAAUGAAUUUAUUUGGCUCAUUUAA-AGCAGCUAAU ......((((((((((....).))))......((((...(((((....))))).))))--.(((.....)))...)))))............(((((((......-.).)))))). ( -31.00) >DroYak_CAF1 67706 110 - 1 A--AGGAGGGUGAUGCGAAAGAUACAUACGACGAAGACGGUGCCU-UUGGCGCACUUC--AGUGGAGCGCAUAAAGCCCUAAAUGAAUUUAUUUGGCUCAUUUAA-AGCAGCUAAU .--...(((((.(((((.........(((...((((...(((((.-..))))).))))--.)))...)))))...)))))............(((((((......-.).)))))). ( -29.00) >DroPer_CAF1 94580 102 - 1 --------------GCCACAGAUACAGCGGCAGGAGUCAGUGCCAUCGGGGGGAUGUCCUUUUGGUGCGCAUUGAGCGGUAAAUGAAUUUAUUUGCCUCAUUUAGUUGUAGCUAAU --------------.....((.((((((.....((.(((((((((((((((((....)))))))))).)))))))..((((((((....)))))))))).....)))))).))... ( -36.80) >consensus A__AAGAGGGUGAUGCGAAAGACAGCGACGACGAAGACGGUGCCUUUUGGCAUACUUC__AGUGGAGCGCAUAAAGCCCUAAAUGAAUUUAUUUGGCUCAUUUAA_AGCAGCUAAU ......(((((.(((((........(.((...((((...(((((....))))).))))...)).)..)))))...)))))............(((((((........).)))))). (-18.38 = -20.38 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:27 2006