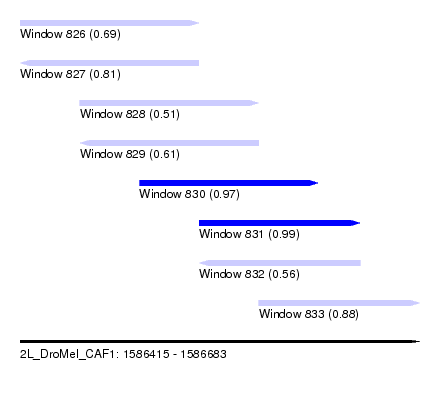
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,586,415 – 1,586,683 |
| Length | 268 |
| Max. P | 0.994888 |

| Location | 1,586,415 – 1,586,535 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -49.94 |
| Consensus MFE | -38.94 |
| Energy contribution | -39.18 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586415 120 + 22407834 UCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGUUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCAUGGUUGGAACCCACUGGUUCUGGA (((((((((((........))))))......((((((((((.(((((.....((((.....))))..))))).)))..)))))))..))))).......(..(((((....)))))..). ( -46.00) >DroSec_CAF1 64938 120 + 1 UCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCUGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGA .(.((..((...((((((((((((.....))))))((((((.((((((..(((((....)))))..)))))).)))..)))))))))....))..)).)(..(((((....)))))..). ( -51.00) >DroSim_CAF1 67245 120 + 1 UCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACCGGUUCUGGA .(.((..((...((((((((((((.....))))))((((((.(((((...(((((....)))))...))))).)))..)))))))))....))..)).)(..(((((....)))))..). ( -49.10) >DroEre_CAF1 66404 120 + 1 UCGGCCGGCACAUCCUUCUGAGCUUUUGAGGCAUCUGCCUCGGGCUCGGAAGGAGGACGCUCCUCGGGAGCCGGAGUUGCGGAUGGAGUUGGUUCGUGGGUGGAAGCCACUGGCUCUGGA ..(((((((((.((((((((((((..((((((....))))))))))))))))))(((((((((((.(.(((....))).).)).)))))..)))))))(((....))).))))))..... ( -60.10) >DroYak_CAF1 65042 114 + 1 UCGGCAGGA------UGUUGUGCUUUCGAGACAUCUGCGUCGGGCUCAGAAUGGGGAUGCUCCUCGGGAGCCGGAGUUGCGGCUGGAGUUGGCUCGUGGGUGGAAGCCACCGGUUCUGGA (((((.(((------((((.((....)).)))))))..)))))..(((((((((((...(..((((.(((((((..((......))..))))))).))))..)...)).)).))))))). ( -43.50) >consensus UCGGCAGGCACAUCCAUUGGUGCUUUUGAGGUAUCUGCCUCGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGA ((((.((((((........))))))))))..((((((((((.(((((...(((((....)))))...))))).)))..)))))))(((..((....(((.......)))))..))).... (-38.94 = -39.18 + 0.24)

| Location | 1,586,415 – 1,586,535 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -39.72 |
| Consensus MFE | -27.44 |
| Energy contribution | -29.68 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586415 120 - 22407834 UCCAGAACCAGUGGGUUCCAACCAUGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAACAUCCUCCUUCUGAGCCCGAGGCAGAUACCUCAAAAGCACCAAUGGAUGUGCCUGCCGA ..(((....(((.(((((.......))))).)))..((((((..(((.(((((..(((((......)))))..))))).))))))))).........((((........))))))).... ( -38.20) >DroSec_CAF1 64938 120 - 1 UCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCAGAGCCCGAGGCAGAUACCUCAAAAGCACCAAUGGAUGUGCCUGCCGA ..(((....(((.(((((.......))))).)))..((((((..(((.(((((..((((((....))))))..))))).))))))))).........((((........))))))).... ( -40.60) >DroSim_CAF1 67245 120 - 1 UCCAGAACCGGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCUGAGCCCGAGGCAGAUACCUCAAAAGCACCAAUGGAUGUGCCUGCCGA ........(((..(((((((.....((......)).((((((..(((.(((((..((((((....))))))..))))).)))))))))................))))...)))..))). ( -43.80) >DroEre_CAF1 66404 120 - 1 UCCAGAGCCAGUGGCUUCCACCCACGAACCAACUCCAUCCGCAACUCCGGCUCCCGAGGAGCGUCCUCCUUCCGAGCCCGAGGCAGAUGCCUCAAAAGCUCAGAAGGAUGUGCCGGCCGA ....(.(((.((((.......))))...............(((..((((((((..((((((....))))))..))))).(((((....)))))............)))..))).)))).. ( -43.00) >DroYak_CAF1 65042 114 - 1 UCCAGAACCGGUGGCUUCCACCCACGAGCCAACUCCAGCCGCAACUCCGGCUCCCGAGGAGCAUCCCCAUUCUGAGCCCGACGCAGAUGUCUCGAAAGCACAACA------UCCUGCCGA ..(((((..((((((((........)))))).(((((((((......))))).....))))....))..)))))........((((((((..(....)....)))------).))))... ( -33.00) >consensus UCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCUGAGCCCGAGGCAGAUACCUCAAAAGCACCAAUGGAUGUGCCUGCCGA ..(((.....((((.......))))...........((((((..(((.(((((..((((((....))))))..))))).))))))))).........((((........))))))).... (-27.44 = -29.68 + 2.24)

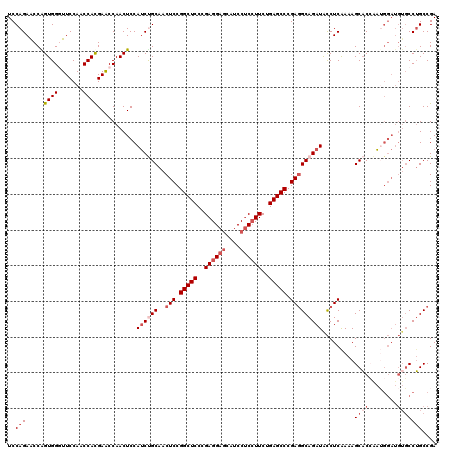
| Location | 1,586,455 – 1,586,575 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -53.90 |
| Consensus MFE | -42.02 |
| Energy contribution | -41.78 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586455 120 + 22407834 CGGGCUCAGAAGGAGGAUGUUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCAUGGUUGGAACCCACUGGUUCUGGAGAUGGUGCGGCAGCCUCCGCAGGAGCUGCUUCUUCUGGGA ....(((((((((((.(.((((((((((.((((.(.((.((((...(((.(((((.......))))).)))...)))).))....).))))....)))).))))))).))))))))))). ( -50.90) >DroSec_CAF1 64978 120 + 1 CGGGCUCUGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGAGUUGGUGCAGCAGCCUCCACAGGAGCUGCUUCUUCUGGGG (.((((((..(((((....)))))..)))))).).....((((.(((((.(((((.((...((((((....))))))(((((((......))).)))).)).)))))))))).))))... ( -51.40) >DroSim_CAF1 67285 120 + 1 CGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACCGGUUCUGGAGUUGGUGCAGCAGCCUCCACAGGAGUUGCUUCUUCUGGGG ....(((((((((((.(.((((((.(((((((((.(((.((((((((.....)))))..))).)))...))))))))(((((((......))).)))).)))))))).))))))))))). ( -49.90) >DroEre_CAF1 66444 120 + 1 CGGGCUCGGAAGGAGGACGCUCCUCGGGAGCCGGAGUUGCGGAUGGAGUUGGUUCGUGGGUGGAAGCCACUGGCUCUGGAGUGGCUGCGCCAGUCUCCGCAGGAGCUGCUUCCUCCGGGG ....((((((.(((((..((((((((((((((((....((((((.......)))))).(((....))).))))))).(((.((((...)))).)))))).))))))..))))))))))). ( -61.10) >DroYak_CAF1 65076 120 + 1 CGGGCUCAGAAUGGGGAUGCUCCUCGGGAGCCGGAGUUGCGGCUGGAGUUGGCUCGUGGGUGGAAGCCACCGGUUCUGGAGUGGCUGCGCCAGUCUCCACAGGAGCUGCUUCCUCCGGCG (.(((((....(((((.....))))).))))).).......(((((((..(((.....(((((...)))))((((((((((((((...))))..))))...)))))))))..))))))). ( -56.20) >consensus CGGGCUCAGAAGGAGGAUGCUCCUCGGGAGCCGGAGUUGCAGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGAGUUGGUGCGGCAGCCUCCACAGGAGCUGCUUCUUCUGGGG ....(((((((((((.(.((((((.(((((((((...........(((....))).(((.......)))))))))))((((.....((....)))))).)))))))).))))))))))). (-42.02 = -41.78 + -0.24)

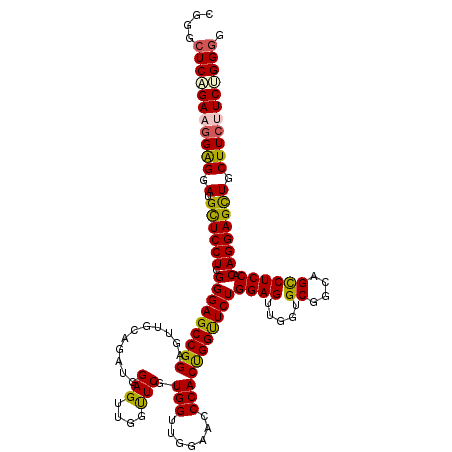
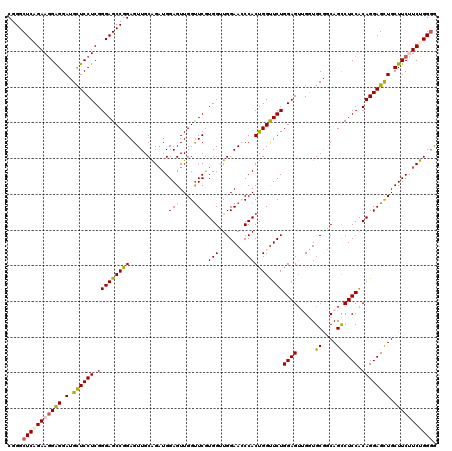
| Location | 1,586,455 – 1,586,575 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -31.00 |
| Energy contribution | -31.12 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586455 120 - 22407834 UCCCAGAAGAAGCAGCUCCUGCGGAGGCUGCCGCACCAUCUCCAGAACCAGUGGGUUCCAACCAUGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAACAUCCUCCUUCUGAGCCCG ...........((((...))))((((((....))........((((...(((.(((((.......))))).)))...))))...))))(((((..(((((......)))))..))))).. ( -36.90) >DroSec_CAF1 64978 120 - 1 CCCCAGAAGAAGCAGCUCCUGUGGAGGCUGCUGCACCAACUCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCAGAGCCCG ........(.(((((((((...))).)))))).)........((((...(((.(((((.......))))).)))...)))).......(((((..((((((....))))))..))))).. ( -39.80) >DroSim_CAF1 67285 120 - 1 CCCCAGAAGAAGCAACUCCUGUGGAGGCUGCUGCACCAACUCCAGAACCGGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCUGAGCCCG ...(((.((......)).))).((((..(((..((((............))))(((((.......)))))..........))).))))(((((..((((((....))))))..))))).. ( -37.90) >DroEre_CAF1 66444 120 - 1 CCCCGGAGGAAGCAGCUCCUGCGGAGACUGGCGCAGCCACUCCAGAGCCAGUGGCUUCCACCCACGAACCAACUCCAUCCGCAACUCCGGCUCCCGAGGAGCGUCCUCCUUCCGAGCCCG ....((((...((.((....))((((((((((..............))))))((.(((.......)))))..))))....))..))))(((((..((((((....))))))..))))).. ( -45.04) >DroYak_CAF1 65076 120 - 1 CGCCGGAGGAAGCAGCUCCUGUGGAGACUGGCGCAGCCACUCCAGAACCGGUGGCUUCCACCCACGAGCCAACUCCAGCCGCAACUCCGGCUCCCGAGGAGCAUCCCCAUUCUGAGCCCG .(((((((......((((((.(((((..((((...)))))))))....(((((((((........)))))).....(((((......))))).))))))))).......))))).))... ( -44.12) >consensus CCCCAGAAGAAGCAGCUCCUGUGGAGGCUGCCGCACCAACUCCAGAACCAGUGGGUUCCAACCACGAACCAACUCCAUCUGCAACUCCGGCUCCCGAGGAGCAUCCUCCUUCUGAGCCCG ...((((....((((...))))((((.(((............))).....((((.......)))).......)))).)))).......(((((..((((((....))))))..))))).. (-31.00 = -31.12 + 0.12)

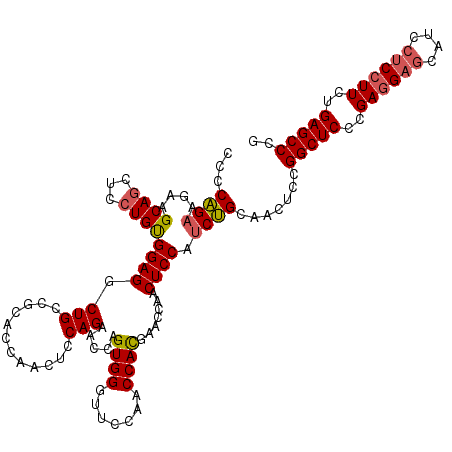
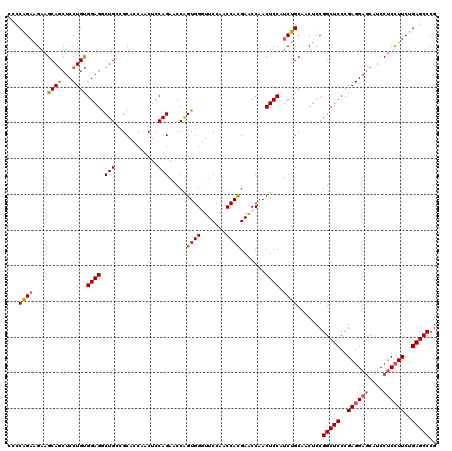
| Location | 1,586,495 – 1,586,615 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.75 |
| Mean single sequence MFE | -60.30 |
| Consensus MFE | -44.06 |
| Energy contribution | -44.38 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586495 120 + 22407834 AGAUGGAGUUGGUUCAUGGUUGGAACCCACUGGUUCUGGAGAUGGUGCGGCAGCCUCCGCAGGAGCUGCUUCUUCUGGGACUGGCUCCGCUGGGGCAGGUUCCUCUGCAGCAGGCUCUGG (((.(((((..((((......((((((....))))))(((((......((((((.(((...))))))))))))))..))))..)))))((((.((.((....)))).))))....))).. ( -49.60) >DroSec_CAF1 65018 120 + 1 AGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGAGUUGGUGCAGCAGCCUCCACAGGAGCUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGG ....(((((..((((.(((..((((((((((..((....))..)))).....(((.((((((((((((((((....))))).)))))))))))))).)))))).)))))))..))))).. ( -61.60) >DroSim_CAF1 67325 120 + 1 AGAUGGAGUUGGUUCGUGGUUGGAACCCACCGGUUCUGGAGUUGGUGCAGCAGCCUCCACAGGAGUUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGG ....(((((..((((.(((..((((((((((..((....))..)))).....(((.((((((((((((((((....))))).)))))))))))))).)))))).)))))))..))))).. ( -60.70) >DroEre_CAF1 66484 120 + 1 GGAUGGAGUUGGUUCGUGGGUGGAAGCCACUGGCUCUGGAGUGGCUGCGCCAGUCUCCGCAGGAGCUGCUUCCUCCGGGGCUGCCUCCUGUGGAGCUGGUUCCUCUGGAGCAGGCUCUGG ....(((((..((((.((((.((((((((((........)))))))..(((((.((((((((((((.(((((....))))).).)))))))))))))))))))))))))))..))))).. ( -62.50) >DroYak_CAF1 65116 120 + 1 GGCUGGAGUUGGCUCGUGGGUGGAAGCCACCGGUUCUGGAGUGGCUGCGCCAGUCUCCACAGGAGCUGCUUCCUCCGGCGCUGCUUCCUGUGGCGCUGGUUCCUCGGGAGCCGGCUCUGG ..(..((((((((((.((((.((((((....((((((((((((((...))))..))))...)))))))))))).(((((((..(.....)..)))))))...)))).))))))))))..) ( -67.10) >consensus AGAUGGAGUUGGUUCGUGGUUGGAACCCACUGGUUCUGGAGUUGGUGCGGCAGCCUCCACAGGAGCUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGG ....(((((..((((...((((..(((.(((........))).))).)))).(((.((((((((((((((((....))))).))))))))))))))...........))))..))))).. (-44.06 = -44.38 + 0.32)

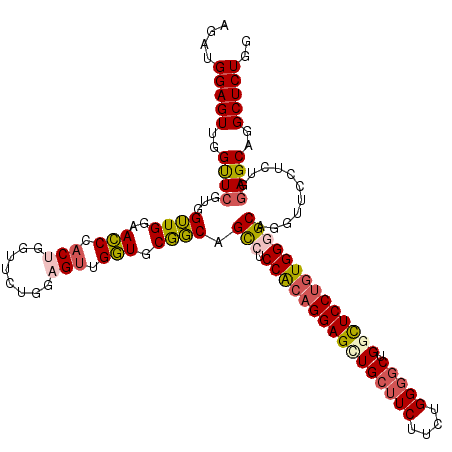
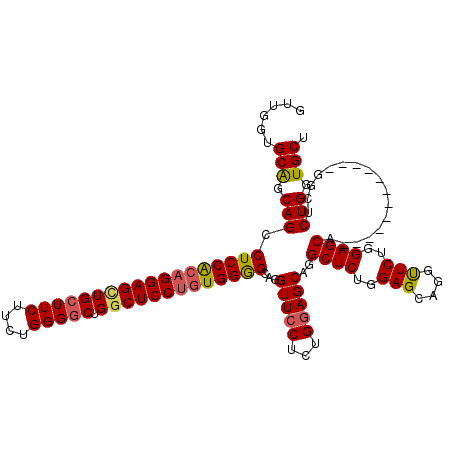
| Location | 1,586,535 – 1,586,643 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -61.98 |
| Consensus MFE | -48.55 |
| Energy contribution | -48.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.06 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586535 108 + 22407834 GAUGGUGCGGCAGCCUCCGCAGGAGCUGCUUCUUCUGGGACUGGCUCCGCUGGGGCAGGUUCCUCUGCAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCU ..((.(((((((((.(((...)))))))))......((((((.(((((...))))).))))))...))).))(((.(..((((..(((....))).------------.))))..).))) ( -49.80) >DroSec_CAF1 65058 108 + 1 GUUGGUGCAGCAGCCUCCACAGGAGCUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCU ........(((((((.((((((((((((((((....))))).))))))))))))))........(..((((..((((..(((....)))..)))).------------.))))..))))) ( -62.50) >DroSim_CAF1 67365 108 + 1 GUUGGUGCAGCAGCCUCCACAGGAGUUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCU ........(((((((.((((((((((((((((....))))).))))))))))))))........(..((((..((((..(((....)))..)))).------------.))))..))))) ( -60.00) >DroEre_CAF1 66524 120 + 1 GUGGCUGCGCCAGUCUCCGCAGGAGCUGCUUCCUCCGGGGCUGCCUCCUGUGGAGCUGGUUCCUCUGGAGCAGGCUCUGGAGCGGGCUCUGGAGCAGGCUCUGGAGCUGGCUCUGGUGCU ....(((((((((.((((((((((((.(((((....))))).).))))))))))))))))(((...)))))))((.(..((((.(((((..(((....)))..))))).))))..).)). ( -75.60) >consensus GUUGGUGCAGCAGCCUCCACAGGAGCUGCUUCUUCUGGGGCUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA____________GGCUCUGGUGCU ......(((.(((.((((((((((((((((((....))))).)))))))))))))...(((((...)))))..((((..(((....)))..)))).................))).))). (-48.55 = -48.80 + 0.25)

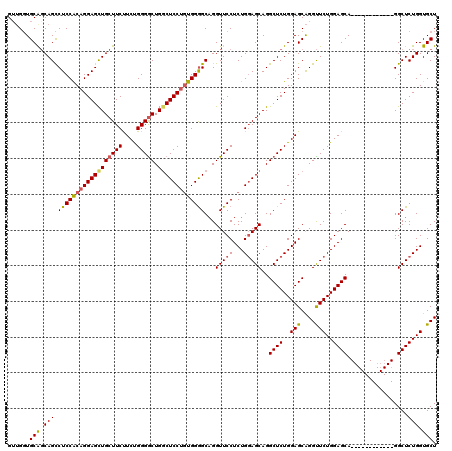
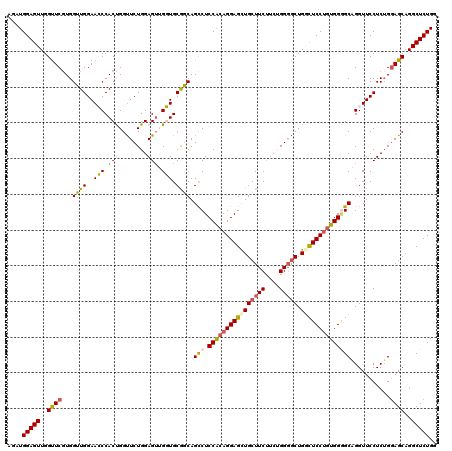
| Location | 1,586,535 – 1,586,643 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -26.48 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586535 108 - 22407834 AGCACCAGAGCC------------UGCUCCAGAACCUGCUCCAGAGCCUGCUGCAGAGGAACCUGCCCCAGCGGAGCCAGUCCCAGAAGAAGCAGCUCCUGCGGAGGCUGCCGCACCAUC .(((.(((.(((------------((...(((...)))...))).))))).)))...((........)).((((((((..((((((.((......)).))).))))))).))))...... ( -36.10) >DroSec_CAF1 65058 108 - 1 AGCACCAGAGCC------------UGCUCCAGAACCUGCUCCAGAGCCUGCUCCAGAGGAACCUGCCCCACAGGAGCCAGCCCCAGAAGAAGCAGCUCCUGUGGAGGCUGCUGCACCAAC .(((.((((((.------------.((((..((......))..))))..))))...........(((((((((((((..((..(....)..)).)))))))))).))))).)))...... ( -42.60) >DroSim_CAF1 67365 108 - 1 AGCACCAGAGCC------------UGCUCCAGAACCUGCUCCAGAGCCUGCUCCAGAGGAACCUGCCCCACAGGAGCCAGCCCCAGAAGAAGCAACUCCUGUGGAGGCUGCUGCACCAAC .(((.((((((.------------.((((..((......))..))))..))))...........((((((((((((...((..(....)..))..))))))))).))))).)))...... ( -39.00) >DroEre_CAF1 66524 120 - 1 AGCACCAGAGCCAGCUCCAGAGCCUGCUCCAGAGCCCGCUCCAGAGCCUGCUCCAGAGGAACCAGCUCCACAGGAGGCAGCCCCGGAGGAAGCAGCUCCUGCGGAGACUGGCGCAGCCAC .((.((.((((..((((..((((..(((....)))..))))..))))..))))....))..((((((((.((((((((..((.....))..))..)))))).)))).)))).))...... ( -52.40) >consensus AGCACCAGAGCC____________UGCUCCAGAACCUGCUCCAGAGCCUGCUCCAGAGGAACCUGCCCCACAGGAGCCAGCCCCAGAAGAAGCAGCUCCUGCGGAGGCUGCCGCACCAAC .((.((...................((((..((......))..))))(((...))).)).....((((((((((((...((..(....)..))..))))))))).)))....))...... (-26.48 = -27.35 + 0.87)

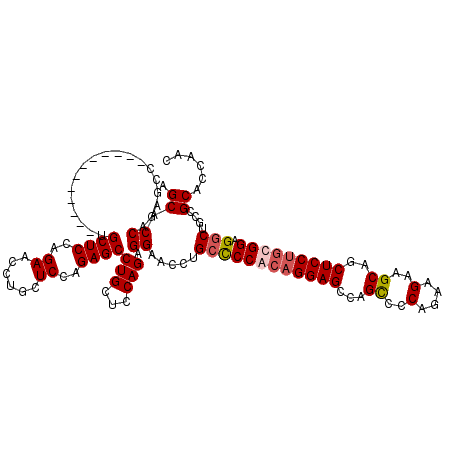
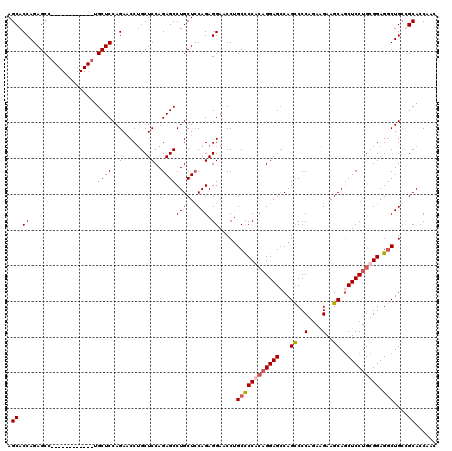
| Location | 1,586,575 – 1,586,683 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -58.06 |
| Consensus MFE | -43.98 |
| Energy contribution | -44.94 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1586575 108 + 22407834 CUGGCUCCGCUGGGGCAGGUUCCUCUGCAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCUGGAUCCGCUGCGGGAGGCUCUUCGACUGGCGCAGGGGGUG ...(((((.(((.(.(((...((((((((((.(((.(..((((..(((....))).------------.))))..).)))(....)))))).)))))........))).).)))))))). ( -50.40) >DroSec_CAF1 65098 108 + 1 CUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCUGGCUCCGCUGCGGGAGGCUCUUCGACUGGCGCAGGGGGUG ...(((((((((((((.(((....(..((((..((((..(((....)))..)))).------------.))))..).))).)))))))((((..((.........))..)))))))))). ( -53.40) >DroSim_CAF1 67405 108 + 1 CUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA------------GGCUCUGGUGCUGGCUCCGCUGCGGGAGGCUCUUCGACUGGCGCAGGUGGUG (((.((((((((((((.(((....(..((((..((((..(((....)))..)))).------------.))))..).))).)))))...)))))))(((........))).)))...... ( -50.60) >DroEre_CAF1 66564 120 + 1 CUGCCUCCUGUGGAGCUGGUUCCUCUGGAGCAGGCUCUGGAGCGGGCUCUGGAGCAGGCUCUGGAGCUGGCUCUGGUGCUGGAUCCGCUGCAGGAGCCUCUUCGACUGGCGCAGGUGGAG (..(((((.((.(((..((((((((((((...(((.(..((((.(((((..(((....)))..))))).))))..).)))...))))..).)))))))..))).)).))...)))..).. ( -65.20) >DroYak_CAF1 65196 120 + 1 CUGCUUCCUGUGGCGCUGGUUCCUCGGGAGCCGGCUCUGGAGCAGGUUCUGGAGCUGGCUCUGGAGCUGGCUCUGGUGCUGGUUCCGCUGCAGGAGCCUCCUCGACUGGCGCAGGUGGAG ((((.(((((..(((((((((((...)))))))))...(((((.(((.(..((((..(((....)))..))))..).))).)))))))..)))))(((.........)))))))...... ( -70.70) >consensus CUGGCUCCUGUGGGGCAGGUUCCUCUGGAGCAGGCUCUGGAGCAGGUUCUGGAGCA____________GGCUCUGGUGCUGGAUCCGCUGCGGGAGGCUCUUCGACUGGCGCAGGUGGUG ..(.((((((((((((..(((((...)))))..)))))(((((.(((.(..((((.((((....)))).))))..).))).)))))...))))))).).(((((.....)).)))..... (-43.98 = -44.94 + 0.96)

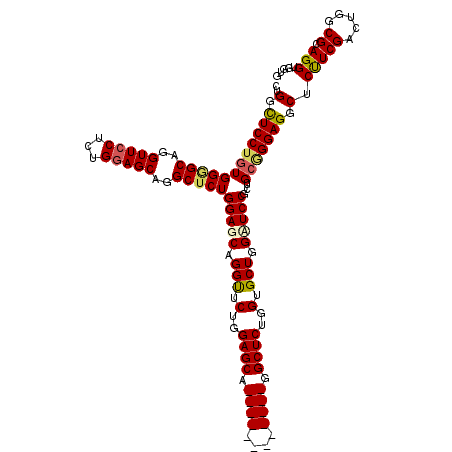
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:13 2006