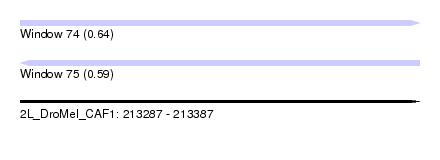
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 213,287 – 213,387 |
| Length | 100 |
| Max. P | 0.638669 |

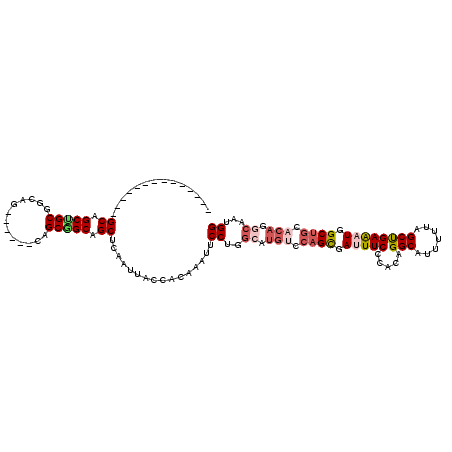
| Location | 213,287 – 213,387 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -38.09 |
| Consensus MFE | -25.80 |
| Energy contribution | -24.75 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
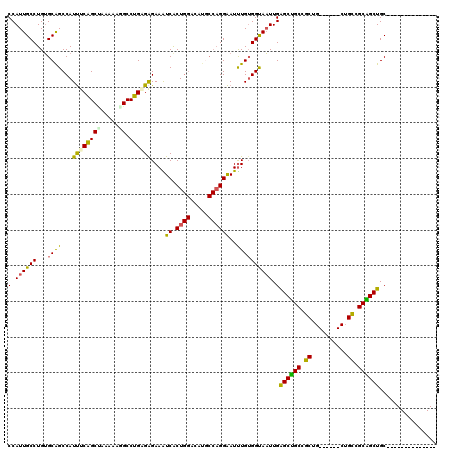
>2L_DroMel_CAF1 213287 100 + 22407834 CCAUUGCCUGUGCAGCCAUUUCAGCUAAAAAGGCCUGUGGGAAAUCGCUGGACAUGCCAGGAAUUUGUGGUAAUUGAGCUGCCGCUG------CUGUCGCAGCUGC-------------- ((((..((((.(((.(((.....(((.....)))..((((....)))))))...))))))).....))))......((((((.((..------..)).))))))..-------------- ( -31.90) >DroSec_CAF1 5585 100 + 1 CCAUUGCCUGUGCAGCCAUUUCAGCUAAAAAUGCCUGGGGGAAAUCGCUGGACAUGCCAGGAAUUUGUGGUAAUUGAGCUGCCGCUG------CUGCAGCGGCUGC-------------- ((((..((((.(((.....((((((........((....)).....))))))..))))))).....)))).......((.(((((((------...))))))).))-------------- ( -34.22) >DroSim_CAF1 5573 100 + 1 CCAUUGCCUGUGCAGCCAUUUCAGCUAAAAAUGCCUGGGGGAAAUCGCUGGACAUGCCAGGAAUUUGUGGUAAUUGAGCUGCCGCUG------CUGCAGCGGCUGC-------------- ((((..((((.(((.....((((((........((....)).....))))))..))))))).....)))).......((.(((((((------...))))))).))-------------- ( -34.22) >DroEre_CAF1 5878 100 + 1 CCAUUGCCUGUGCAGCCAUUUCGGCUAAAAAGGCCUGUGAGAAAUCACUGGACAUCCCAGGAAUUUGUGGUAAUUGAGCUGCUGCUG------CUGCCGCAGCUGC-------------- ((((..((((...((((.....)))).....(.((.((((....)))).)).)....)))).....))))......((((((.((..------..)).))))))..-------------- ( -35.50) >DroYak_CAF1 7633 97 + 1 CCAUUGCCUGUGCAGCCAUUUCAGCCAAAAAGGCCUGAGAGAAGUCACU---------AGGAAUUUGUGGCAAUUGAGCUGCCGCAGCAGCCGCAGCCGCAGCUGC-------------- ...((((.((((((((((((((((((.....)).))))))...(((((.---------........)))))...)).)))).))))))))..(((((....)))))-------------- ( -35.60) >DroMoj_CAF1 6059 114 + 1 CCAUUGCCUGGGCGGCCAUCUCGGCGAGAAGUGCCUGAGAGAAGUCACUGGACAUGCCUGGGAUCUGAGGCAUUUGGGCAGCUGCGG------CUGCGGCUGCUGCGGCGGCAGCUGCUG (((.(((((.((...((.((((((((.....)))).))))...(((....))).......))..)).)))))..)))((((((((.(------((((((...))))))).)))))))).. ( -57.10) >consensus CCAUUGCCUGUGCAGCCAUUUCAGCUAAAAAGGCCUGAGAGAAAUCACUGGACAUGCCAGGAAUUUGUGGUAAUUGAGCUGCCGCUG______CUGCCGCAGCUGC______________ (.((((((...((((...((((((((.....)).))))))....((.((((.....))))))..)))))))))).)((((((.((..........)).))))))................ (-25.80 = -24.75 + -1.05)

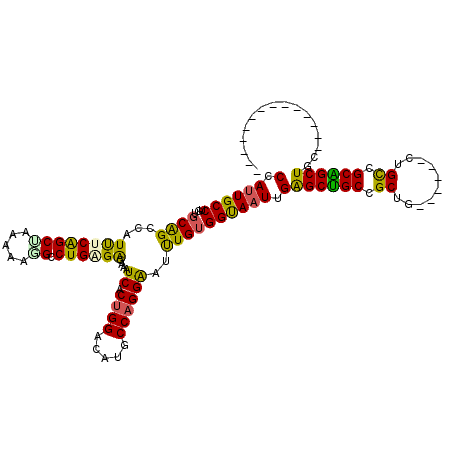
| Location | 213,287 – 213,387 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -35.25 |
| Consensus MFE | -21.99 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 213287 100 - 22407834 --------------GCAGCUGCGACAG------CAGCGGCAGCUCAAUUACCACAAAUUCCUGGCAUGUCCAGCGAUUUCCCACAGGCCUUUUUAGCUGAAAUGGCUGCACAGGCAAUGG --------------((.(((((....)------)))).))...................((..((.(((.((((.(((((.....(((.......)))))))).)))).))).))...)) ( -30.70) >DroSec_CAF1 5585 100 - 1 --------------GCAGCCGCUGCAG------CAGCGGCAGCUCAAUUACCACAAAUUCCUGGCAUGUCCAGCGAUUUCCCCCAGGCAUUUUUAGCUGAAAUGGCUGCACAGGCAAUGG --------------((.(((((((...------))))))).))................((..((.(((.((((.(((((.....(((.......)))))))).)))).))).))...)) ( -33.40) >DroSim_CAF1 5573 100 - 1 --------------GCAGCCGCUGCAG------CAGCGGCAGCUCAAUUACCACAAAUUCCUGGCAUGUCCAGCGAUUUCCCCCAGGCAUUUUUAGCUGAAAUGGCUGCACAGGCAAUGG --------------((.(((((((...------))))))).))................((..((.(((.((((.(((((.....(((.......)))))))).)))).))).))...)) ( -33.40) >DroEre_CAF1 5878 100 - 1 --------------GCAGCUGCGGCAG------CAGCAGCAGCUCAAUUACCACAAAUUCCUGGGAUGUCCAGUGAUUUCUCACAGGCCUUUUUAGCCGAAAUGGCUGCACAGGCAAUGG --------------(.((((((.((..------..)).))))))).....(((.((((..((((.....))))..)))).......((((.(.(((((.....))))).).))))..))) ( -33.10) >DroYak_CAF1 7633 97 - 1 --------------GCAGCUGCGGCUGCGGCUGCUGCGGCAGCUCAAUUGCCACAAAUUCCU---------AGUGACUUCUCUCAGGCCUUUUUGGCUGAAAUGGCUGCACAGGCAAUGG --------------((((((((....))))))))(((.((((((.....((((.(((..(((---------((.(....).)).)))..))).))))......))))))....))).... ( -37.40) >DroMoj_CAF1 6059 114 - 1 CAGCAGCUGCCGCCGCAGCAGCCGCAG------CCGCAGCUGCCCAAAUGCCUCAGAUCCCAGGCAUGUCCAGUGACUUCUCUCAGGCACUUCUCGCCGAGAUGGCCGCCCAGGCAAUGG ..((((((((.((.((.......)).)------).))))))))(((.((((((........))))))(((..(((.((..((((.(((.......))))))).)).)))...)))..))) ( -43.50) >consensus ______________GCAGCUGCGGCAG______CAGCGGCAGCUCAAUUACCACAAAUUCCUGGCAUGUCCAGCGAUUUCCCACAGGCAUUUUUAGCUGAAAUGGCUGCACAGGCAAUGG ..............((.(((((.............))))).))................((..((.(((.((((.(((((.....(((.......)))))))).)))).))).))...)) (-21.99 = -23.10 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:22 2006