| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,710,629 – 14,710,758 |
| Length | 129 |
| Max. P | 0.998099 |

| Location | 14,710,629 – 14,710,725 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14710629 96 + 22407834 UGAAAUUCAUAUGGAUGGAAAU--GGAACCACCCACACGCCACCCCACAUAUUCAGCUCCAGGAAUUUGCACCCACCUGCA--GGUGGGGCAUAACAUUC ..((((((...(((((((...(--((......)))....)))...............)))).))))))((.((((((....--))))))))......... ( -26.56) >DroSec_CAF1 67393 96 + 1 UGAAAUUCAUAUGGAUGGAAAU--GGAACCACCCGCACGCCGCCCCACAUAUUCAGCUCCCGGAAUUUGCACCCAACUGCA--GGUGGGGCAUAACAUAC .........((((...((...(--((......)))....))(((((((..((((........))))(((((......))))--))))))))....)))). ( -25.70) >DroSim_CAF1 67617 96 + 1 UGAAAUUCAUAUGGAUGGAAAU--GGAACCACCCGCACGCCGCCCCACAAAUUCAGCUCCCGGAAUUUGCACCCAACUGCA--GGUGGGGCAUAACAUAC .........((((...((...(--((......)))....))(((((((((((((........))))))(((......))).--.)))))))....)))). ( -26.70) >DroEre_CAF1 66847 76 + 1 UGAAAUUCAUAUGGAUGGAAAU--GGCAACACCCACAUGCCGCCCCACCUAUG--------------------CACCUGCA--GGUGGGGCAUAAGAUAC ............(((((....(--(....))....))).))(((((((((..(--------------------(....)))--))))))))......... ( -27.60) >DroYak_CAF1 66634 81 + 1 UGAAAUUCAUGUGGAUGGAUAU--GGAGCCACCCACUCGCCGCCCCACAAAUUCAGC--------------CCCACCUGCA---GUGGGGCAUAAAAUAC .........(((((.(((...(--((......)))....)))..)))))......((--------------(((((.....---)))))))......... ( -26.60) >DroPer_CAF1 93443 89 + 1 UGAAAUUCAUAUGGAUGGGAAGACAGACAUAUCUACACACAACCCCCCUUC--UAACCCC-----UUUU----UACCUGGAUAAGGGUGGCAUAAAAUAC ........(((((..((......))..)))))..........((.((((((--((.....-----....----....))))..)))).)).......... ( -13.52) >consensus UGAAAUUCAUAUGGAUGGAAAU__GGAACCACCCACACGCCGCCCCACAUAUUCAGCUCC_____UUUG__CCCACCUGCA__GGUGGGGCAUAACAUAC .....(((((....))))).....((......)).......(((((((....................................)))))))......... (-12.96 = -12.77 + -0.19)

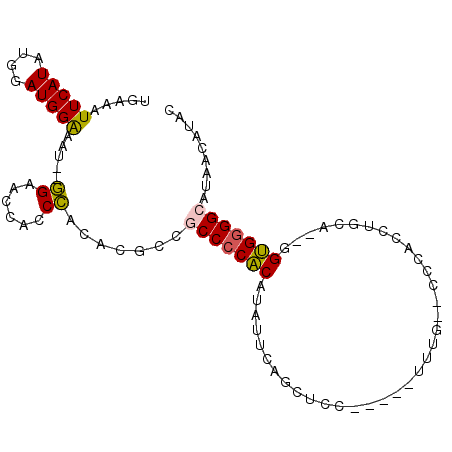

| Location | 14,710,629 – 14,710,725 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -22.25 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14710629 96 - 22407834 GAAUGUUAUGCCCCACC--UGCAGGUGGGUGCAAAUUCCUGGAGCUGAAUAUGUGGGGUGGCGUGUGGGUGGUUCC--AUUUCCAUCCAUAUGAAUUUCA ((((....(((((((((--....)))))).))).))))...............((..((..(((((((((((....--....))))))))))).))..)) ( -37.20) >DroSec_CAF1 67393 96 - 1 GUAUGUUAUGCCCCACC--UGCAGUUGGGUGCAAAUUCCGGGAGCUGAAUAUGUGGGGCGGCGUGCGGGUGGUUCC--AUUUCCAUCCAUAUGAAUUUCA ((((((..((((((((.--..((((((((.......))))...)))).....))))))))))))))((((((....--....))))))............ ( -36.90) >DroSim_CAF1 67617 96 - 1 GUAUGUUAUGCCCCACC--UGCAGUUGGGUGCAAAUUCCGGGAGCUGAAUUUGUGGGGCGGCGUGCGGGUGGUUCC--AUUUCCAUCCAUAUGAAUUUCA ((((((..(((((((((--(......))))((((((((.(....).))))))))))))))))))))((((((....--....))))))............ ( -39.50) >DroEre_CAF1 66847 76 - 1 GUAUCUUAUGCCCCACC--UGCAGGUG--------------------CAUAGGUGGGGCGGCAUGUGGGUGUUGCC--AUUUCCAUCCAUAUGAAUUUCA ........(((((((((--(((....)--------------------)..)))))))))).((((((((((..(..--..)..))))))))))....... ( -33.20) >DroYak_CAF1 66634 81 - 1 GUAUUUUAUGCCCCAC---UGCAGGUGGG--------------GCUGAAUUUGUGGGGCGGCGAGUGGGUGGCUCC--AUAUCCAUCCACAUGAAUUUCA (...((((((((((((---..(((.....--------------.))).....))))))).....((((((((....--....)))))))))))))...). ( -30.90) >DroPer_CAF1 93443 89 - 1 GUAUUUUAUGCCACCCUUAUCCAGGUA----AAAA-----GGGGUUA--GAAGGGGGGUUGUGUGUAGAUAUGUCUGUCUUCCCAUCCAUAUGAAUUUCA (...((((((((.(((((((((.....----....-----.))))..--.)))))))..((..((.(((((....)))))...))..))))))))...). ( -16.10) >consensus GUAUGUUAUGCCCCACC__UGCAGGUGGG__CAAA_____GGAGCUGAAUAUGUGGGGCGGCGUGUGGGUGGUUCC__AUUUCCAUCCAUAUGAAUUUCA .........(((((((.....(((....................))).....)))))))..(((((((((((..........)))))))))))....... (-22.25 = -23.08 + 0.84)

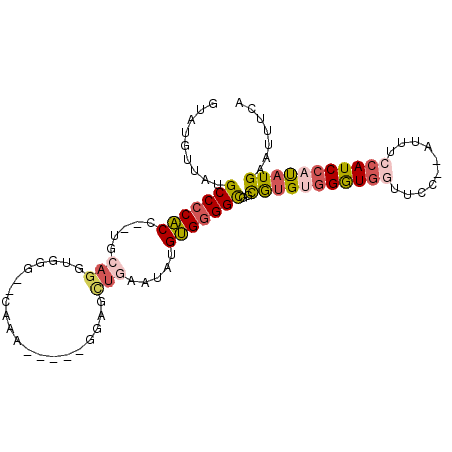

| Location | 14,710,667 – 14,710,758 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.32 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14710667 91 + 22407834 CACCCCACAUAUUCAGCUCCAGGAAUUUGCACCCACCUGCA--GGUGGGGCAUAACAUUCUGACUGGGCUAAUGCAUUGCU-------AGCCACCCAAAU .............(((...(((((...(((.((((((....--))))))))).....))))).)))(((((..((...)))-------))))........ ( -27.40) >DroPse_CAF1 94452 89 + 1 AACCCCCCUUC--UAACCCC-----UUUU----UACCUGGAUAAGGGUGGCAUAAAAUACAGCUACGGCUAAUGCAUUGCCUUUCAGAAGGAACCCUAAU ......(((((--(...(((-----((.(----(.....)).))))).((((........(((....))).......))))....))))))......... ( -23.46) >DroSec_CAF1 67431 91 + 1 CGCCCCACAUAUUCAGCUCCCGGAAUUUGCACCCAACUGCA--GGUGGGGCAUAACAUACUGACUGGGCUAAUGCAUUGCU-------AGCCACCCAAAU .(((((((..((((........))))(((((......))))--))))))))...............(((((..((...)))-------))))........ ( -26.40) >DroSim_CAF1 67655 91 + 1 CGCCCCACAAAUUCAGCUCCCGGAAUUUGCACCCAACUGCA--GGUGGGGCAUAACAUACUGACUGGGCUAAUGCAUUGCU-------AGCCACCUAAAU .(((((((((((((........))))))(((......))).--.)))))))...............(((((..((...)))-------))))........ ( -27.40) >DroEre_CAF1 66885 71 + 1 CGCCCCACCUAUG--------------------CACCUGCA--GGUGGGGCAUAAGAUACUGCCAAGGCUAAUGCAUUGCU-------AGCCACCCAAAU .(((((((((..(--------------------(....)))--))))))))...............(((((..((...)))-------))))........ ( -27.70) >DroYak_CAF1 66672 76 + 1 CGCCCCACAAAUUCAGC--------------CCCACCUGCA---GUGGGGCAUAAAAUACUGCCUGGGCUAAUGCAUUGCC-------AGCCACCCAAAU .(((((((.....(((.--------------.....)))..---))))))).........((.((((((....))....))-------)).))....... ( -20.90) >consensus CGCCCCACAUAUUCAGCUCC_____UUUG__CCCACCUGCA__GGUGGGGCAUAACAUACUGACUGGGCUAAUGCAUUGCU_______AGCCACCCAAAU .(((((((....................................)))))))...............(((.........)))................... (-12.32 = -12.32 + 0.00)

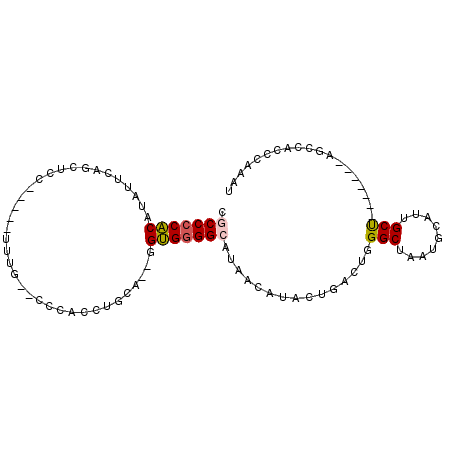
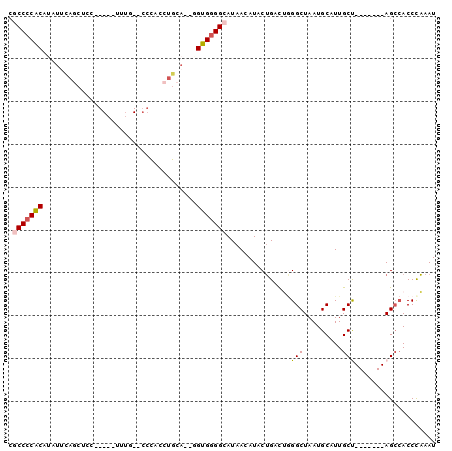
| Location | 14,710,667 – 14,710,758 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.96 |
| Mean single sequence MFE | -28.89 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.29 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14710667 91 - 22407834 AUUUGGGUGGCU-------AGCAAUGCAUUAGCCCAGUCAGAAUGUUAUGCCCCACC--UGCAGGUGGGUGCAAAUUCCUGGAGCUGAAUAUGUGGGGUG ............-------.((..(((((((((((((...((((....(((((((((--....)))))).))).)))))))).)))))...))))..)). ( -31.00) >DroPse_CAF1 94452 89 - 1 AUUAGGGUUCCUUCUGAAAGGCAAUGCAUUAGCCGUAGCUGUAUUUUAUGCCACCCUUAUCCAGGUA----AAAA-----GGGGUUA--GAAGGGGGGUU .......((((((((((..((((......((((....)))).......)))).(((((.........----..))-----))).)))--))))))).... ( -25.42) >DroSec_CAF1 67431 91 - 1 AUUUGGGUGGCU-------AGCAAUGCAUUAGCCCAGUCAGUAUGUUAUGCCCCACC--UGCAGUUGGGUGCAAAUUCCGGGAGCUGAAUAUGUGGGGCG ..((((((.((.-------......))....))))))............(((((((.--..((((((((.......))))...)))).....))))))). ( -30.80) >DroSim_CAF1 67655 91 - 1 AUUUAGGUGGCU-------AGCAAUGCAUUAGCCCAGUCAGUAUGUUAUGCCCCACC--UGCAGUUGGGUGCAAAUUCCGGGAGCUGAAUUUGUGGGGCG ......(.((((-------((.......)))))))..............((((((((--(......))))((((((((.(....).))))))))))))). ( -29.70) >DroEre_CAF1 66885 71 - 1 AUUUGGGUGGCU-------AGCAAUGCAUUAGCCUUGGCAGUAUCUUAUGCCCCACC--UGCAGGUG--------------------CAUAGGUGGGGCG ........((((-------((.......))))))...............((((((((--(((....)--------------------)..))))))))). ( -27.10) >DroYak_CAF1 66672 76 - 1 AUUUGGGUGGCU-------GGCAAUGCAUUAGCCCAGGCAGUAUUUUAUGCCCCAC---UGCAGGUGGG--------------GCUGAAUUUGUGGGGCG .........((.-------......))....((((..((((.((((...(((((((---.....)))))--------------)).)))))))).)))). ( -29.30) >consensus AUUUGGGUGGCU_______AGCAAUGCAUUAGCCCAGGCAGUAUGUUAUGCCCCACC__UGCAGGUGGG__CAAA_____GGAGCUGAAUAUGUGGGGCG ..((((((.((..............))....))))))............(((((((.....(((....................))).....))))))). (-17.84 = -18.29 + 0.45)

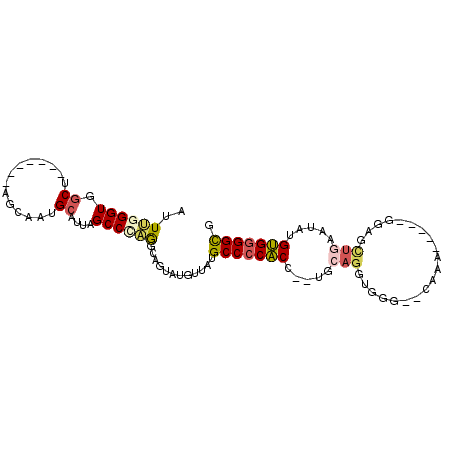
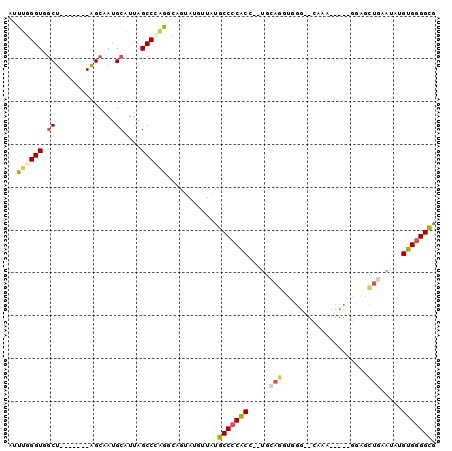
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:25 2006