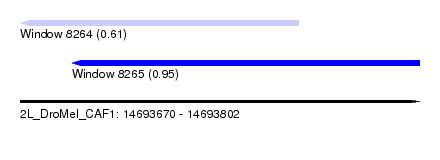
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,693,670 – 14,693,802 |
| Length | 132 |
| Max. P | 0.947921 |

| Location | 14,693,670 – 14,693,762 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.53 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -21.78 |
| Energy contribution | -21.12 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
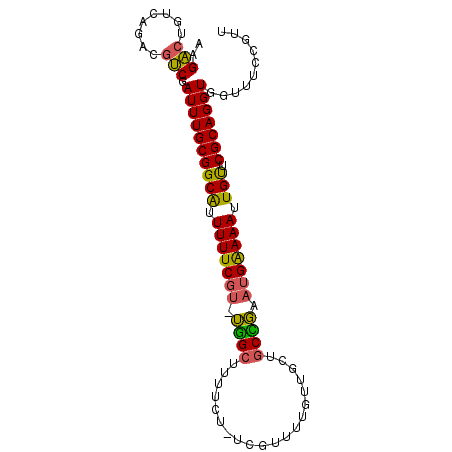
>2L_DroMel_CAF1 14693670 92 - 22407834 AAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UCGUUUUGUUGCUGCCGAAUGAAAAUUGUUCGCAGGUGGUUUCCGUU ..........((((.(.((((((((((.(((((((-((((....(.-.......).....))).)))))))).)).)))))))).)....)))) ( -24.30) >DroSim_CAF1 50083 92 - 1 AAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UCGUUUUGUUGCUGCCGAAUGAAAAUUGUUCGCAGGUGGUUUCCGUU ..........((((.(.((((((((((.(((((((-((((....(.-.......).....))).)))))))).)).)))))))).)....)))) ( -24.30) >DroEre_CAF1 49490 92 - 1 AAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGUUUGGCUUUUCU-UCGUU-UGCUGCUGCCGAAUGAAAAUUGUUCGCAGGUGGUUUCCGUU ..........((((.(.((((((((((.((((((((((((......-.....-.......)))))))))))).)).)))))))).)....)))) ( -28.67) >DroYak_CAF1 48145 92 - 1 AAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UCGUUUUGUUGCUGCCGAAUGAAAAUUGUUCGCAGGUGGUUUCCGUU ..........((((.(.((((((((((.(((((((-((((....(.-.......).....))).)))))))).)).)))))))).)....)))) ( -24.30) >DroMoj_CAF1 56841 85 - 1 UAGACUGUCAGCCGUCGAUUUGCGGCGUUUUUCAC-GAGUUUUUUUUUCGAGU--------CUUAUGGGAAAUUGUUCGCAGGUGGCUGCCGUC ..(((.(.(((((....(((((((((((((..((.-.((.(((......))).--------))..))..))).)).)))))))))))))).))) ( -25.70) >DroAna_CAF1 47215 92 - 1 AAGCCCGACAGUCUGCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UGUAUCUGAAGCUGCUGAAUGAAAAUUGCUCGCAGGUGGUUUCCGUU ......(((.(...((.((((((((((.(((((((-((((.((((.-.......))))..))).)))))))).))).))))))).))..).))) ( -28.70) >consensus AAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU_UGGCUUUUCU_UCGUUUUGUUGCUGCCGAAUGAAAAUUGUUCGCAGGUGGUUUCCGUU ..(((........))).((((((((((.(((((((.((((....................)))).))))))).))).))))))).......... (-21.78 = -21.12 + -0.66)


| Location | 14,693,687 – 14,693,802 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.74 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -23.96 |
| Energy contribution | -23.02 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
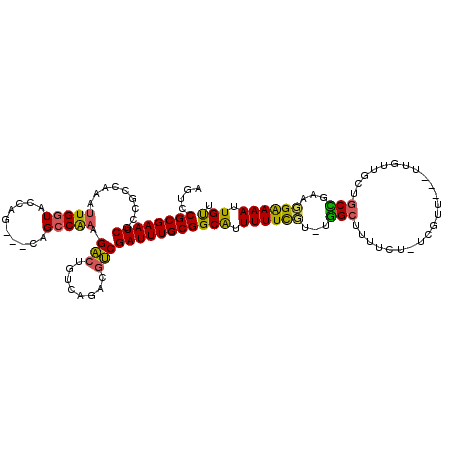
>2L_DroMel_CAF1 14693687 115 - 22407834 AGUCCGCGAAUCGCUGCCAAAUUGGUACCAGUACCAGCCAAAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UCGUU---UUGUUGCUGCCGAAUGAAAAUUGUU ...(((((((((((((....((((....))))..))))....(((........))))))))))))((.(((((((-((((....(.-.....---..).....))).)))))))).)).. ( -33.20) >DroPse_CAF1 63341 111 - 1 AGUCCGCGAAUCGCCGCCAAAUUGGUACCAGA---CGCCGAAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCCU-UGGCUUUU---UCGUUGCCUGUUUUUUGCCU--GGAAAAUUGUU ...(((((((((...(((.....)))....((---(((.((......)).).)))))))))))))((.((((((.-.(((....---................))).--)))))).)).. ( -32.35) >DroEre_CAF1 49507 115 - 1 AGUCCGCGAAUCGCUGCCAAAUUGGUACCAGUACCAGCCAAAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGUUUGGCUUUUCU-UCGUU----UGCUGCUGCCGAAUGAAAAUUGUU ...(((((((((((((....((((....))))..))))....(((........))))))))))))((.((((((((((((......-.....----.......)))))))))))).)).. ( -37.57) >DroMoj_CAF1 56858 102 - 1 AGCGCGCGAAUCGACGGCAAAUUGGUAC------CAGCCAUAGACUGUCAGCCGUCGAUUUGCGGCGUUUUUCAC-GAGUUUUUUUUUCGAG---U--------CUUAUGGGAAAUUGUU .((.((((((((((((((....((((..------..))))..(.....).))))))))))))))))((((..((.-.((.(((......)))---.--------))..))..)))).... ( -39.20) >DroAna_CAF1 47232 109 - 1 AGUCCGCGAAUCGCCGCCAAAUUGGUAC------CAGGCGAAGCCCGACAGUCUGCGAUUUGCGGCAUUUUUCGU-UGGCUUUUCU-UGUAU---CUGAAGCUGCUGAAUGAAAAUUGCU ....((((((((((.(((.....)))..------.((((...........))))))))))))))(((.(((((((-((((.((((.-.....---..))))..))).)))))))).))). ( -36.40) >DroPer_CAF1 71335 111 - 1 AGUCCGCGAAUCGCCGCCAAAUUGGUACCAGA---CGCCGAAGACUGUCAGACGUCGAUUUGCGGCAUUUUUUCU-UGGCUUUU---UCGUUGCCUGUUUUUUGCCU--GGAAAAUUGUU .(.(((((((((...(((.....)))....((---(((.((......)).).))))))))))))))...((((((-.(((....---................))).--))))))..... ( -30.85) >consensus AGUCCGCGAAUCGCCGCCAAAUUGGUACCAG___CAGCCAAAGACUGUCAGACGUCGAUUUGCGGCAUUUUUCGU_UGGCUUUUCU_UCGUU___UUGUUGCUGCCGAAGGAAAAUUGUU ....((((((((.........(((((..........))))).(((........)))))))))))(((.((((((...(((.......................)))...)))))).))). (-23.96 = -23.02 + -0.94)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:16 2006