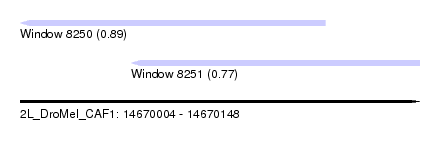
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,670,004 – 14,670,148 |
| Length | 144 |
| Max. P | 0.890983 |

| Location | 14,670,004 – 14,670,114 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -16.33 |
| Energy contribution | -16.24 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
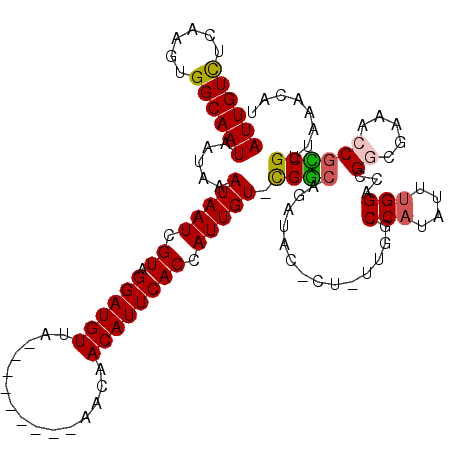
>2L_DroMel_CAF1 14670004 110 - 22407834 AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA---------AACAACAUUCACCAUUGU-CGGCAGAUACGUUUUUGGCCAUAUUUGGAUCGGCGAAACCGCCGUUAAACAU ...(((.((((((((........(((((.((.(((((((.---------...))))))))).)))))-...((((......)))))))))..))))))(((((....)))))........ ( -26.40) >DroPse_CAF1 35168 106 - 1 AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUGAACAAAAACUAACAACAUUCACCAUUGUGCGACA------------ACCAUAUUUGGGCCGCCU-GCCCGUCGG-ACAAAU .((((((((.(((((............(((((.((((((((...............)))).)))).)))))..------------.(((....)))))))).)-)......))-)))).. ( -27.26) >DroEre_CAF1 24557 110 - 1 AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUUGGAUGUUA---------AACAACAUUCACCAUUGU-UGGCAUAUACUCUUUUGGCCAUAUUUGGAGCGGCGAAACCGCCGUUAAACAU ((((((......)))))).............((((((((.---------...))))))))(((....-((((.((........))))))....)))(((((((....)))))))...... ( -30.40) >DroYak_CAF1 24921 110 - 1 AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA---------AACAACAUUCACCAUUGU-UGGCAGAUACUCUUUUGGCCAUAUUUGGAGCGGCGAAACCGCCGUUAAACAU ((((((......))))))..............(((((((.---------...))))))).(((....-(((((((......))).))))....)))(((((((....)))))))...... ( -31.10) >DroAna_CAF1 24186 109 - 1 AUUGUUUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUAA---------AACAACAUUCACCAUUGU-UGGCAGAUAUGCU-CUGGCCAUAUUGGGCUCGGCGGAACCACCGUUAGACAU ..(((((((((((((.......((((((.((.((((((..---------....)))))))).)))))-)((((....))))-...))))).))))....(((((.....))))).)))). ( -26.40) >DroPer_CAF1 38394 106 - 1 AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUGAACAAAAACUAACAACAUUCACCAUUGUGCGGCA------------ACCAUAUUUGGGCCGCCU-GCCCGUCGG-ACAAAU .((((((...(.((((.......(((((.((.((((((...............)))))))).)))))(((((.------------.(((....)))))))).)-))))...))-)))).. ( -31.16) >consensus AUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA_________AACAACAUUCACCAUUGU_CGGCAGAUAC_CU_UUGGCCAUAUUUGGACCGGCGAAACCGCCGUUAAACAU ((((((......)))))).....(((((.((.((((((...............)))))))).))))).((((..............(((....)))...((.....))))))........ (-16.33 = -16.24 + -0.08)

| Location | 14,670,044 – 14,670,148 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.20 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -15.53 |
| Energy contribution | -15.39 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
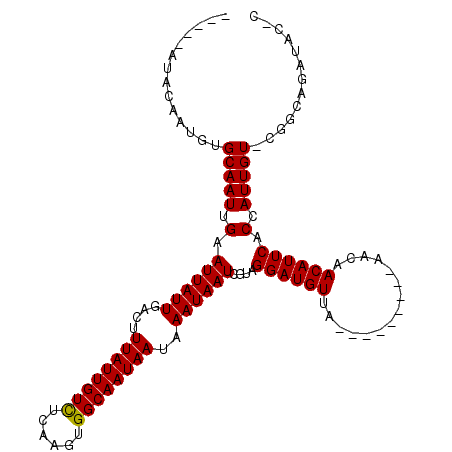
>2L_DroMel_CAF1 14670044 104 - 22407834 ACAAUAUACAAUGUGCAAUUGAAUUAUUGACUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA---------AACAACAUUCACCAUUGU-CGGCAGAUACGU .(((((..((((.....))))...)))))...((((((((......))))))))........(((((((((((.---------...))))))).((.....-.))....)))). ( -20.40) >DroPse_CAF1 35201 102 - 1 -----GUGUGUGUUGCAAUUGAAUUAUUGAGUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUGAACAAAAACUAACAACAUUCACCAUUGUGCGACA------- -----.....(((((((..((.((((((....((((((((......))))))))..))))))....((((((...............))))))..))...)))))))------- ( -23.26) >DroEre_CAF1 24597 104 - 1 ACAAUAUACAAUGUGCAAUUGAAUUAUUGACUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUUGGAUGUUA---------AACAACAUUCACCAUUGU-UGGCAUAUACUC ..........((((((....((.(((((....((((((((......))))))))..)))))))(.((((((((.---------...)))))))).).....-..)))))).... ( -23.30) >DroYak_CAF1 24961 99 - 1 -----AUACAAUGUGCAAUUGAAUUAUUGACUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA---------AACAACAUUCACCAUUGU-UGGCAGAUACUC -----.....((.(((................((((((((......))))))))..((((((.((.(((((((.---------...))))))))).)))))-).))).)).... ( -20.80) >DroAna_CAF1 24225 102 - 1 --CAUGUACCAUGUGCAAUUGAAUUAUUGAGUUUAUUGUUUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUAA---------AACAACAUUCACCAUUGU-UGGCAGAUAUGC --(((((.(((...(((((.(.((((((....((((((((......))))))))..))))))....((((((..---------....)))))).).)))))-)))...))))). ( -19.90) >DroPer_CAF1 38427 100 - 1 -------GUGUGUUGCAAUUGAAUUAUUGAGUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUGAACAAAAACUAACAACAUUCACCAUUGUGCGGCA------- -------...(((((((..((.((((((....((((((((......))))))))..))))))....((((((...............))))))..))...)))))))------- ( -22.76) >consensus _____AUACAAUGUGCAAUUGAAUUAUUGACUUUAUUGUCUCAAGUGGCAAUAAUAAAUAAUCGUAGGAUGUUA_________AACAACAUUCACCAUUGU_CGGCAGAUAC_C ..............(((((.(.((((((....((((((((......))))))))..))))))....((((((...............)))))).).)))))............. (-15.53 = -15.39 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:38:03 2006