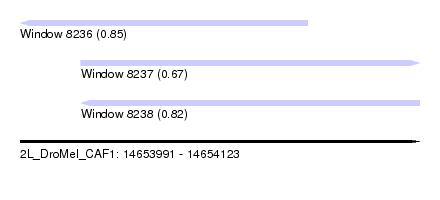
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,653,991 – 14,654,123 |
| Length | 132 |
| Max. P | 0.853564 |

| Location | 14,653,991 – 14,654,086 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 93.36 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.59 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
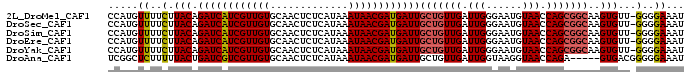
>2L_DroMel_CAF1 14653991 95 - 22407834 CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU-GGGGAAAU ....(((((((((((((((((((((((.............))))))))))))(((((((.(((......))).)))))))...)).)-)))))))) ( -28.62) >DroSec_CAF1 8793 95 - 1 CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU-GGGGAAAU ....(((((((((((((((((((((((.............))))))))))))(((((((.(((......))).)))))))...)).)-)))))))) ( -28.62) >DroSim_CAF1 10090 95 - 1 CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU-GGGGAAAU ....(((((((((((((((((((((((.............))))))))))))(((((((.(((......))).)))))))...)).)-)))))))) ( -28.62) >DroEre_CAF1 8610 95 - 1 CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU-GGGGAAAU ....(((((((((((((((((((((((.............))))))))))))(((((((.(((......))).)))))))...)).)-)))))))) ( -28.62) >DroYak_CAF1 9078 95 - 1 CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU-GGGGAAAU ....(((((((((((((((((((((((.............))))))))))))(((((((.(((......))).)))))))...)).)-)))))))) ( -28.62) >DroAna_CAF1 8469 91 - 1 UCGGCUCUUUUUACUGAUCGUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGUAAGGUAACCAGA-----GUGACGGGGGAAAU ....((((..(((((((((((((((((.............))))))))))))........(((((......))))))-----))))..)))).... ( -25.72) >consensus CCAUGUUUUCUUACAGAUCAUCGUUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCAGCGGCAAGUGUU_GGGGAAAU .....((..(.(((.((((((((((((.............))))))))))))(((((((.(((......))).)))))))..)))...)..))... (-23.33 = -23.59 + 0.25)
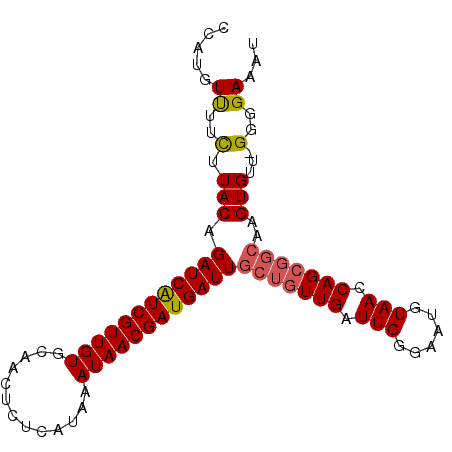
| Location | 14,654,011 – 14,654,123 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.668009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14654011 112 + 22407834 UGGUUACAUUCCCAAUCAACAGCAAUCAUCGUUAUUUAUGAGAGUUGCACAA--CGAUGAUCUGUAAGAAAACAUGGAAAAAAAUCGCAGACCCAAAAUAAAGGGGACCAAAAG--- (((((...((((...((.((((..(((((((((....((....)).....))--)))))))))))..))......))))............(((........))))))))....--- ( -24.50) >DroPse_CAF1 12243 104 + 1 GGGUUACAUUACCAAUCAACAGCAAUCAUCGUUAUUUAUGAGA-UUGCACAACACGAUGAUCGGUCUGAGAGGCCCGAAA-AAC-----GAC--GAAAAAAUGGU--CCU--GGGGU ......((..((((.......((((((.((((.....))))))-))))......((.(..((((.((....)).))))..-).)-----)..--.......))))--..)--).... ( -24.50) >DroEre_CAF1 8630 111 + 1 UGGUUACAUUCCCAAUCAACAGCAAUCAUCGUUAUUUAUGAGAGUUGCACAA--CGAUGAUCUGUAAGAAAACAUGGAAA-AAAUGCAAGACCCAAAAUAAAGGGUACCAGAAG--- ((((....((((...((.((((..(((((((((....((....)).....))--)))))))))))..))......)))).-.........((((........))))))))....--- ( -23.40) >DroYak_CAF1 9098 111 + 1 UGGUUACAUUCCCAAUCAACAGCAAUCAUCGUUAUUUAUGAGAGUUGCACAA--CGAUGAUCUGUAAGAAAACAUGGAAA-AAAUGCAAGACCCAAAAUAAAGGGGACCAGAAG--- (((((...((((...((.((((..(((((((((....((....)).....))--)))))))))))..))......)))).-..........(((........))))))))....--- ( -24.50) >DroAna_CAF1 8485 109 + 1 UGGUUACCUUACCAAUCAACAGCAAUCAUCGUUAUUUAUGAGAGUUGCACAA--CGACGAUCAGUAAAAAGAGCCGAAAAAAUAAGCAAAACCCAGGAACAUGGGGCCCA---G--- .((((...((((..(((..(.(((((..((((.....))))..)))))....--.)..)))..))))....))))................((((......)))).....---.--- ( -20.20) >DroPer_CAF1 15640 103 + 1 GGGUUACAUUACCAAUCAACAGCAAUCAUCGUUAUUUAUGAGA-UUGCACAACACGAUGAUCGGUCUGAGAGGCCCGAAA-AAC-----GAC--GAAAAAAUGGUAACCC--AG--- (((((((..............((((((.((((.....))))))-))))......((.(..((((.((....)).))))..-).)-----)..--.........)))))))--..--- ( -31.20) >consensus UGGUUACAUUACCAAUCAACAGCAAUCAUCGUUAUUUAUGAGAGUUGCACAA__CGAUGAUCUGUAAGAAAACACGGAAA_AAAUGCAAGACCCAAAAAAAAGGGGACCA__AG___ (((((.....(((........(((((..((((.....))))..)))))............((((..........))))........................))))))))....... (-13.06 = -13.08 + 0.03)

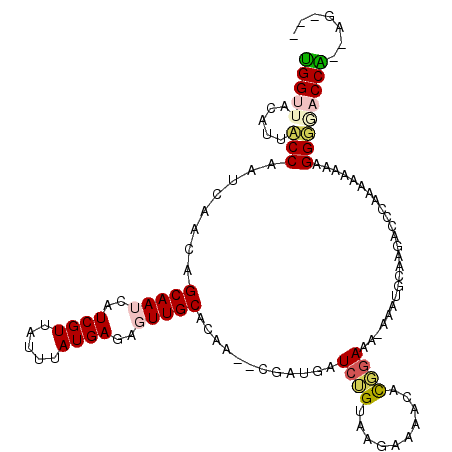
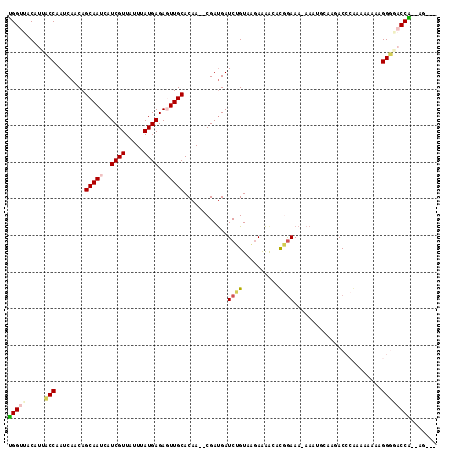
| Location | 14,654,011 – 14,654,123 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -12.85 |
| Energy contribution | -12.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14654011 112 - 22407834 ---CUUUUGGUCCCCUUUAUUUUGGGUCUGCGAUUUUUUUCCAUGUUUUCUUACAGAUCAUCG--UUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCA ---....((((.(((........)))...........((((((.....((..(((((((((((--((((.............)))))))))))..)))).)).))))))....)))) ( -27.02) >DroPse_CAF1 12243 104 - 1 ACCCC--AGG--ACCAUUUUUUC--GUC-----GUU-UUUCGGGCCUCUCAGACCGAUCAUCGUGUUGUGCAA-UCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGUAAUGUAACCC .((..--(((--((.((......--)).-----)))-))..))(((..((((..((((((((((.(((((...-...)))))...))))))))))...))))..))).......... ( -26.20) >DroEre_CAF1 8630 111 - 1 ---CUUCUGGUACCCUUUAUUUUGGGUCUUGCAUUU-UUUCCAUGUUUUCUUACAGAUCAUCG--UUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCA ---.....(((((((........))))..(((((..-.(((((.....((..(((((((((((--((((.............)))))))))))..)))).)).))))))))))))). ( -30.42) >DroYak_CAF1 9098 111 - 1 ---CUUCUGGUCCCCUUUAUUUUGGGUCUUGCAUUU-UUUCCAUGUUUUCUUACAGAUCAUCG--UUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCA ---.....(((.(((........)))...(((((..-.(((((.....((..(((((((((((--((((.............)))))))))))..)))).)).))))))))))))). ( -28.72) >DroAna_CAF1 8485 109 - 1 ---C---UGGGCCCCAUGUUCCUGGGUUUUGCUUAUUUUUUCGGCUCUUUUUACUGAUCGUCG--UUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGUAAGGUAACCA ---.---.((..((((......))))(((((((.(((....((((..........((((((((--((((.............))))))))))))))))..))).)))))))...)). ( -27.02) >DroPer_CAF1 15640 103 - 1 ---CU--GGGUUACCAUUUUUUC--GUC-----GUU-UUUCGGGCCUCUCAGACCGAUCAUCGUGUUGUGCAA-UCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGUAAUGUAACCC ---..--(((((((.........--.((-----(..-...)))(((..((((..((((((((((.(((((...-...)))))...))))))))))...))))..)))...))))))) ( -33.40) >consensus ___CU__UGGUCCCCAUUAUUUUGGGUCUUGCAUUU_UUUCCAGCUUUUCUUACAGAUCAUCG__UUGUGCAACUCUCAUAAAUAACGAUGAUUGCUGUUGAUUGGGAAUGUAACCA ............(((.......((((.............))))............((((((((..(((((.......)))))....))))))))..........))).......... (-12.85 = -12.65 + -0.19)

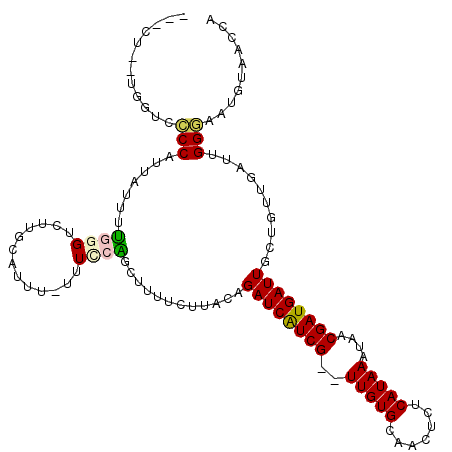
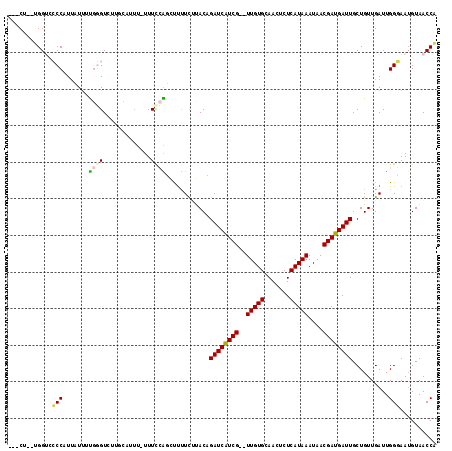
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:51 2006