
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
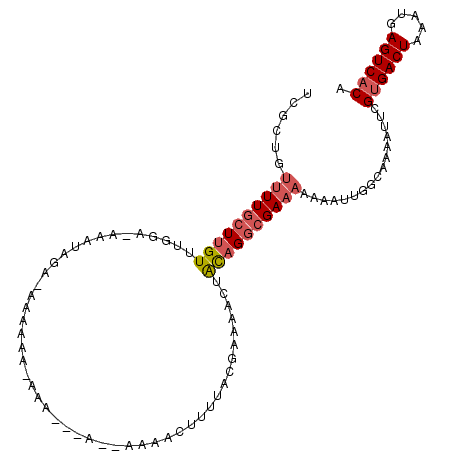
| Location | 14,638,412 – 14,638,547 |
| Length | 135 |
| Max. P | 0.986160 |

| Location | 14,638,412 – 14,638,509 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.95 |
| Mean single sequence MFE | -19.34 |
| Consensus MFE | -11.89 |
| Energy contribution | -12.61 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
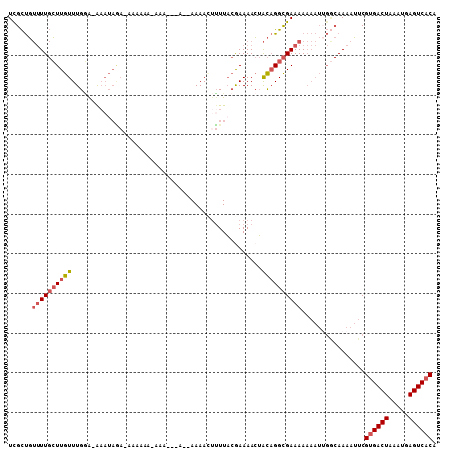
>2L_DroMel_CAF1 14638412 97 + 22407834 UCGCUGUUUUGCUUGUUUGGG-G----GA-AAAAAAAAUA---A--AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACA ..(((.((((((((((((.(.-.----((-(.........---.--.....)))..).))....))))))))))......))).......((((((.....)))))). ( -19.36) >DroSec_CAF1 2330 103 + 1 UCGCUGUUUUGCUUGUUUGGA-AAAUAGA-AAAAAAUAAA---AAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGCGACUAAAUGAGUCACA ..(((.((((((((((.....-...((((-(.........---........)))))........))))))))))......))).......(.((((.....)))).). ( -15.32) >DroSim_CAF1 3629 101 + 1 UUGCUGUUUUGCUUGUUUGGA-AAAUAGA-AAAAAAUAAA---A--AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACA (((((.((((((((((.....-...((((-(.........---.--.....)))))........))))))))))......))))).....((((((.....)))))). ( -20.65) >DroEre_CAF1 1918 90 + 1 UCGCUGUUUUGCUUGUUUGGA-AAAUAGA-----------------AAAAAUUUUACAAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACA ..(((.(((((((((((((.(-((((...-----------------....))))).))).....))))))))))......))).......((((((.....)))))). ( -20.70) >DroYak_CAF1 2358 91 + 1 UUGCUGUUUUGCUUGUUUGAA-AAAUAAA---------U-------AAAAAAUUUACGAAAACUACAGGCGAAAAAAAUGGACAAAAUUCGUGACUAAAUGAGUCACA (((((.((((((((((.....-...((((---------(-------.....)))))........)))))))))).....)).))).....((((((.....)))))). ( -18.99) >DroPer_CAF1 6543 96 + 1 UCGUCUGGUU-GUUGUUUGCACAAAUAC-CAAAAAAAACACCGA--AAAAUUGUUUAAAAAAAAGUCGGUGAAAA--------AAAAAUCGUGACUAAAUGAGUCACA .....((((.-.((((....))))..))-)).......((((((--....((.......))....))))))....--------.......((((((.....)))))). ( -21.00) >consensus UCGCUGUUUUGCUUGUUUGGA_AAAUAGA_AAAAAA_AAA___A__AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACA ......((((((((((................................................))))))))))................((((((.....)))))). (-11.89 = -12.61 + 0.72)

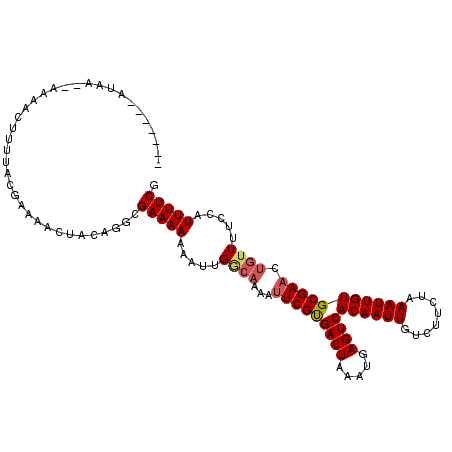
| Location | 14,638,436 – 14,638,547 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -18.86 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
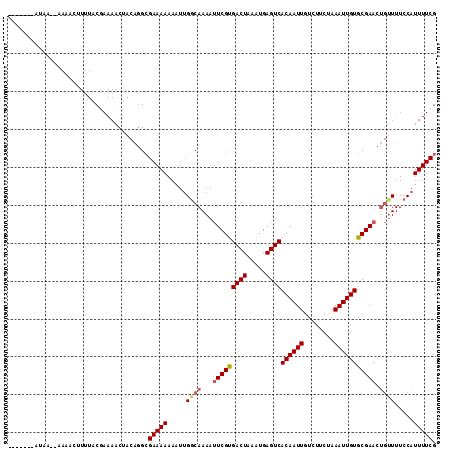
>2L_DroMel_CAF1 14638436 111 + 22407834 AAAAAAAAUAA--AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG ...........--..........((((((..((((((.((.....)).))....(((((((((.....))))((((((........)))))))))))))))......)))))) ( -19.50) >DroSec_CAF1 2358 113 + 1 AAAAAAUAAAAAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGCGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG .......................((((((..((((((.((.....)).))....(((((((((.....))))((((((........)))))))))))))))......)))))) ( -22.00) >DroSim_CAF1 3657 111 + 1 AAAAAAUAAAA--AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG ...........--..........((((((..((((((.((.....)).))....(((((((((.....))))((((((........)))))))))))))))......)))))) ( -19.50) >DroEre_CAF1 1946 100 + 1 -------------AAAAAUUUUACAAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG -------------.......................((((((....(((.(((.(((((((((.....))))((((((........)))))))))))...))).))))))))) ( -17.10) >DroYak_CAF1 2386 101 + 1 --------U----AAAAAAUUUACGAAAACUACAGGCGAAAAAAAUGGACAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG --------.----...........((((((.....(((........((((((.....((((((.....))))))..)))))).........))).....))))))........ ( -19.53) >DroAna_CAF1 2222 85 + 1 -------AUAA--AAAAUUUAUACAAAAAGUACUCGGGAAAA-------------UCGUGACUAAAUGAGUCACAAUUGUCUUCCAAAUUGUGCGAACUGA------UUUUCG -------....--................((((...((((((-------------(.((((((.....)))))).)))...)))).....))))(((....------..))). ( -15.50) >consensus _______AUAA__AAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAACUGUUUUCCAUUUUCG .....................................(((((.....((((...(((((((((.....))))((((((........))))))))))).)))).....))))). (-14.57 = -15.03 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:46 2006