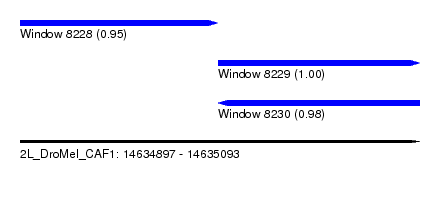
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,634,897 – 14,635,093 |
| Length | 196 |
| Max. P | 0.999441 |

| Location | 14,634,897 – 14,634,994 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -29.79 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
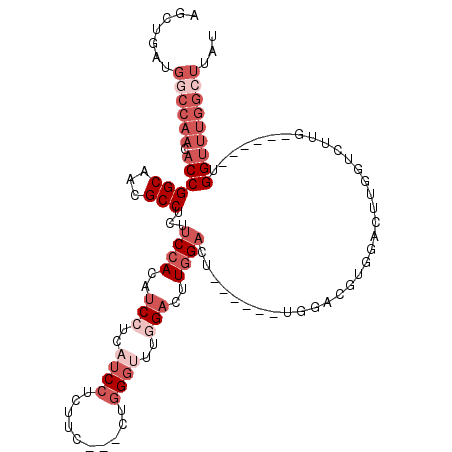
>2L_DroMel_CAF1 14634897 97 + 22407834 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUUGCAGCCCCCACCAGUCCC (((((...((...(((((((...((((((.((...(((.(((........))))))..)).))))))))))))).))..)))))............. ( -33.20) >DroSec_CAF1 26546 93 + 1 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUCGCAGCCCCCA---GU-CC ((((..((.((.((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).)))))).)).))...))---))-.. ( -34.10) >DroSim_CAF1 16993 93 + 1 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUCGCAACCCCCA---GU-CC ((((..((.((.((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).)))))).)).))...))---))-.. ( -33.80) >DroEre_CAF1 17117 93 + 1 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUCGAAGCCCCCA---AA-AG ...(((.(((..((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).)))))))))))).....---..-.. ( -31.30) >DroYak_CAF1 18047 93 + 1 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACGAAUCGCCACGAGAGAGUGAAGUGACGACGUCAAAGCCCCCA---GU-CC ...((((..((.((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).))))))....))...))---))-.. ( -30.00) >DroAna_CAF1 15644 93 + 1 GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUCGCAGCCUCCA---GC-CG ((((..((.((.((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).)))))).)).))...))---))-.. ( -36.20) >consensus GCUGCUGUCGCCACGUCGCUACGUCAUUCGCUAAUUGGUCGAGAUACAAAUCGCCACGAGAGAGUGAAGUGACGACGUCGCAGCCCCCA___GU_CC ...(((((....((((((.(((.((((((.((...(((.(((........))))))..)).)))))).))).)))))).)))))............. (-29.79 = -30.15 + 0.36)


| Location | 14,634,994 – 14,635,093 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 68.22 |
| Mean single sequence MFE | -35.24 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.64 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.39 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14634994 99 + 22407834 AUAAGCCAAACC--------AAGACCAAGUCCACAUCCA------AGUCCAAGUCCAAACCCAG---GAAGAGGAUGAGGAUGUGGAAGAGGCGUUGCCGGUGUUGGCCAUCAGCU ...(((....((--------((.(((...(((((((((.------.((((...(((.......)---))...))))..)))))))))...(((...)))))).))))......))) ( -39.50) >DroPse_CAF1 24590 83 + 1 ----------------------GGCUAUGGCUAUGGCUA------UUGCUAUGGCUAUGGCUAUGGCUAUGAGGAAGAGGAUGUGGAAGAGGCGUUGCC-----UGGACGACAACG ----------------------(((((((((((((((((------(....)))))))))))))))))).............(((.(...((((...)))-----)...).)))... ( -32.80) >DroSec_CAF1 26639 95 + 1 AUAAGCCAAACCAUAAGACCAAGACCAA------------------GUCCAAGUCCAAACCCAG---GAAGAGGAUGAGGAUGUGGAAGAGGCGUUGCCGGUGUUGGCCAUCAGCU ....((((((((......(((...((..------------------((((...(((.......)---))...))))..))...)))....(((...)))))).)))))........ ( -28.10) >DroSim_CAF1 17086 107 + 1 AUAAGCCAAACCAUAAGACCAAGACCAAGUCGACGUCCA------AGUCCAAGUCCAAACCCAG---GAAGAGGAUGAGGAUGUGGAAGAGGCGUUGCCGGUGUUGGCCAUCAGCU ................(.((((.(((...((.((((((.------.((((...(((.......)---))...))))..)))))).))...(((...)))))).)))).)....... ( -34.40) >DroYak_CAF1 18140 110 + 1 AUAAGCCAGUCCA---AGGCAAAGCCAAGUCCACGUCCAAGACCAAGUCCAAGUCUAAGCCCAG---GAAGAGGAUGAGGAUGUGGAAGAGGCGUUGCCGGUGUUGGUCACCAGCU ...(((.......---.(((((.(((...(((((((((..(.((...(((..((....))...)---))...)).)..)))))))))...))).)))))((((.....)))).))) ( -41.40) >consensus AUAAGCCAAACCA______CAAGACCAAGUCCACGUCCA______AGUCCAAGUCCAAACCCAG___GAAGAGGAUGAGGAUGUGGAAGAGGCGUUGCCGGUGUUGGCCAUCAGCU ....((((((((...................................((((.((((....((..........))....)))).))))...(((...)))))).)))))........ (-13.84 = -14.64 + 0.80)

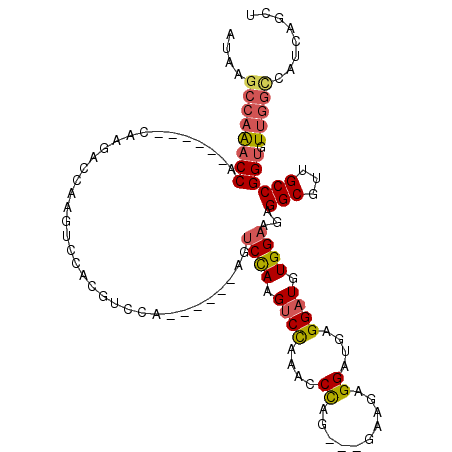
| Location | 14,634,994 – 14,635,093 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 68.22 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -9.56 |
| Energy contribution | -13.16 |
| Covariance contribution | 3.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14634994 99 - 22407834 AGCUGAUGGCCAACACCGGCAACGCCUCUUCCACAUCCUCAUCCUCUUC---CUGGGUUUGGACUUGGACU------UGGAUGUGGACUUGGUCUU--------GGUUUGGCUUAU (((..(..(((((.((((((...)))...(((((((((...(((...((---(.......)))...)))..------.)))))))))...))).))--------))))..)))... ( -39.60) >DroPse_CAF1 24590 83 - 1 CGUUGUCGUCCA-----GGCAACGCCUCUUCCACAUCCUCUUCCUCAUAGCCAUAGCCAUAGCCAUAGCAA------UAGCCAUAGCCAUAGCC---------------------- (((((((.....-----)))))))...............................((.((.((.((.((..------..)).)).)).)).)).---------------------- ( -13.50) >DroSec_CAF1 26639 95 - 1 AGCUGAUGGCCAACACCGGCAACGCCUCUUCCACAUCCUCAUCCUCUUC---CUGGGUUUGGACUUGGAC------------------UUGGUCUUGGUCUUAUGGUUUGGCUUAU (((..(.((((((.((((((...)))...((((..(((..((((.....---..))))..)))..)))).------------------..))).)))))).......)..)))... ( -30.30) >DroSim_CAF1 17086 107 - 1 AGCUGAUGGCCAACACCGGCAACGCCUCUUCCACAUCCUCAUCCUCUUC---CUGGGUUUGGACUUGGACU------UGGACGUCGACUUGGUCUUGGUCUUAUGGUUUGGCUUAU (((..(.((((((.((((((.(((((...((((..(((..((((.....---..))))..)))..))))..------.)).))).).).)))).)))))).......)..)))... ( -34.20) >DroYak_CAF1 18140 110 - 1 AGCUGGUGACCAACACCGGCAACGCCUCUUCCACAUCCUCAUCCUCUUC---CUGGGCUUAGACUUGGACUUGGUCUUGGACGUGGACUUGGCUUUGCCU---UGGACUGGCUUAU (((..((..((((....(((((.(((...(((((.(((....((...((---(.((.......)).)))...))....))).)))))...))).))))))---)))))..)))... ( -41.90) >consensus AGCUGAUGGCCAACACCGGCAACGCCUCUUCCACAUCCUCAUCCUCUUC___CUGGGUUUGGACUUGGACU______UGGACGUGGACUUGGUCUUG______UGGUUUGGCUUAU .......((((((.((((((...)))...((((..(((..((((..........))))..)))..))))...................................)))))))))... ( -9.56 = -13.16 + 3.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:43 2006