
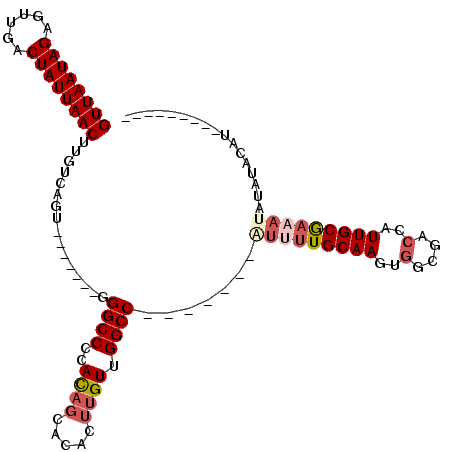
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,628,778 – 14,628,869 |
| Length | 91 |
| Max. P | 0.879337 |

| Location | 14,628,778 – 14,628,869 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -18.19 |
| Energy contribution | -19.33 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14628778 91 + 22407834 GUUAAUAGAGUUGACUAUUAACUUGUCAGU-------GGGCCCCAUAGCACACUUGUUGGCC-------AUUUUGCAAGUGGCGACUAUUGCAAAAUAUAUACAU--------- ((((((((......))))))))......((-------(((((....((((....))))))))-------(((((((((..(....)..)))))))))...)))..--------- ( -24.50) >DroSec_CAF1 10954 91 + 1 GUUAAUAGAGUUGACUAUUAACUUGUCAGU-------GGGCCCCACAGCACACUUGUUGGCC-------AUUUUGCAAGUGGCGACCAUUGCGAAAUAUAUACAU--------- ((((((((......))))))))(((.((((-------((......(((((....)))))(((-------(((.....))))))..)))))))))...........--------- ( -27.00) >DroSim_CAF1 10604 100 + 1 GUUAAUAGAGUUGACUAUUAACUUGUCAGU-------GGGCCCCACAGCACACUUGUUGGCC-------AUUUUGCAAGUGGCGACCAUUGCGAAAUAUAUACAUACUUUAGCC ((((((((......))))))))(((.((((-------((......(((((....)))))(((-------(((.....))))))..))))))))).................... ( -27.00) >DroEre_CAF1 10852 87 + 1 GUUAAUAGAGUUGACUAUUAACUUGUCAGU-------GGGCCCCACAGCACACUUGUUGGCC-------AUUUUGCAAGUGGCAACCAUUGCG------CGAAAUGA------- ((((((((......))))))))((((((((-------((......(((((....)))))(((-------(((.....))))))..)))))).)------))).....------- ( -27.60) >DroYak_CAF1 11636 100 + 1 GUUAAUAGAGUUGACUAUUAACUUGUCAGUGGGCAGUGGGCCCCACAGCUCACUUGUUGGCC-------GUUUUGCAAGUGGCGAGCAUUGCGACAUAUCUACAUAC------- ((((((((......)))))))).(((((((((((.((((...)))).)))))))((((.(((-------(.........)))).))))....))))...........------- ( -33.80) >DroAna_CAF1 9993 86 + 1 GUUAAUAGAGUUAACUAUUAACUUGUCAGU-------GGGCCCCACAGC------GUUGGCCGUUUUCCGUUUUGCAAGAGGCGCCCAUUGCGAAAGGC--------------- ((((((((......))))))))(((.((((-------((((((....((------(.(((.......)))...)))....)).))))))))))).....--------------- ( -27.50) >consensus GUUAAUAGAGUUGACUAUUAACUUGUCAGU_______GGGCCCCACAGCACACUUGUUGGCC_______AUUUUGCAAGUGGCGACCAUUGCGAAAUAUAUACAU_________ ((((((((......))))))))................((((..((((.....)))).)))).......(((((((((..(....)..)))))))))................. (-18.19 = -19.33 + 1.14)

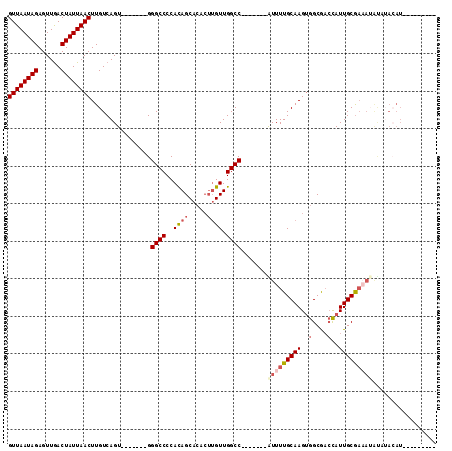
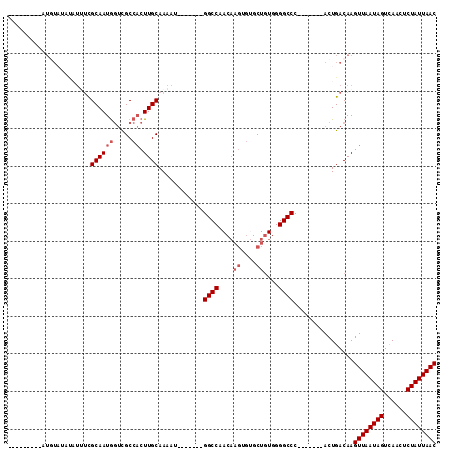
| Location | 14,628,778 – 14,628,869 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -16.38 |
| Energy contribution | -17.05 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14628778 91 - 22407834 ---------AUGUAUAUAUUUUGCAAUAGUCGCCACUUGCAAAAU-------GGCCAACAAGUGUGCUAUGGGGCCC-------ACUGACAAGUUAAUAGUCAACUCUAUUAAC ---------.(((...(((((((((..(((....)))))))))))-------)....)))((((.(((....))).)-------))).....((((((((......)))))))) ( -22.30) >DroSec_CAF1 10954 91 - 1 ---------AUGUAUAUAUUUCGCAAUGGUCGCCACUUGCAAAAU-------GGCCAACAAGUGUGCUGUGGGGCCC-------ACUGACAAGUUAAUAGUCAACUCUAUUAAC ---------.............(((.(((.(.((((..(((...(-------(.....))....))).)))).).))-------).)).)..((((((((......)))))))) ( -22.60) >DroSim_CAF1 10604 100 - 1 GGCUAAAGUAUGUAUAUAUUUCGCAAUGGUCGCCACUUGCAAAAU-------GGCCAACAAGUGUGCUGUGGGGCCC-------ACUGACAAGUUAAUAGUCAACUCUAUUAAC (((((((((((....)))))).(((((((...))).))))....)-------))))....((((.(((....))).)-------))).....((((((((......)))))))) ( -25.40) >DroEre_CAF1 10852 87 - 1 -------UCAUUUCG------CGCAAUGGUUGCCACUUGCAAAAU-------GGCCAACAAGUGUGCUGUGGGGCCC-------ACUGACAAGUUAAUAGUCAACUCUAUUAAC -------.......(------((((...(((((((.((....)))-------)).))))...)))))(((.((....-------.)).))).((((((((......)))))))) ( -24.70) >DroYak_CAF1 11636 100 - 1 -------GUAUGUAGAUAUGUCGCAAUGCUCGCCACUUGCAAAAC-------GGCCAACAAGUGAGCUGUGGGGCCCACUGCCCACUGACAAGUUAAUAGUCAACUCUAUUAAC -------...........((((.....((((((.....((.....-------.))......)))))).(((((........))))).)))).((((((((......)))))))) ( -27.70) >DroAna_CAF1 9993 86 - 1 ---------------GCCUUUCGCAAUGGGCGCCUCUUGCAAAACGGAAAACGGCCAAC------GCUGUGGGGCCC-------ACUGACAAGUUAAUAGUUAACUCUAUUAAC ---------------.......(((.(((((.(((((........)))..(((((....------))))))).))))-------).)).)..((((((((......)))))))) ( -26.10) >consensus _________AUGUAUAUAUUUCGCAAUGGUCGCCACUUGCAAAAU_______GGCCAACAAGUGUGCUGUGGGGCCC_______ACUGACAAGUUAAUAGUCAACUCUAUUAAC ......................((((((.....)).))))............((((....((....))....))))................((((((((......)))))))) (-16.38 = -17.05 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:40 2006