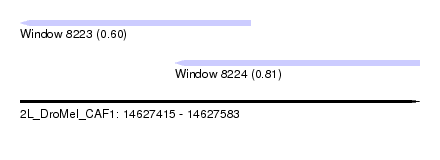
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,627,415 – 14,627,583 |
| Length | 168 |
| Max. P | 0.814550 |

| Location | 14,627,415 – 14,627,512 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -19.57 |
| Energy contribution | -20.90 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14627415 97 - 22407834 --UAUCCCAGCGAGAGCAGAGC------AGGGAAACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG-CCAGCCAGCUCUUUGC --.......((((((((..(((------(((....))))))((.....(((((....)))))..(((((((((........))))))))-)..))..)))).)))) ( -36.80) >DroSim_CAF1 9273 97 - 1 --UAUCCCAGCGAGAGCAGAGC------AGGGAAACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG-CCAGCCAGCUCUUUGC --.......((((((((..(((------(((....))))))((.....(((((....)))))..(((((((((........))))))))-)..))..)))).)))) ( -36.80) >DroEre_CAF1 9462 96 - 1 --UAUCCCAGCGAGAGCAGAGC------AGGU-AACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG-CCAGCCAGCUCUUUGC --.......((((((((..(((------(((.-..))))))((.....(((((....)))))..(((((((((........))))))))-)..))..)))).)))) ( -31.30) >DroWil_CAF1 21292 105 - 1 GUGAGACGAGUGAGAUGGGAGUUAGAAGAAGCUAACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUGGCCAGAAGGUU-UUUGG ......(((((((((((((.(((((......)))))...)).)))).....))))))).......((((((((........)))))))).(((((.....-))))) ( -27.10) >DroYak_CAF1 10287 96 - 1 --UAUCCCAGCGAGAGCAGAGC------AGGG-AACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG-CCAGCCAGCUCUUCUC --.........((((((..(((------(((.-..))))))((.....(((((....)))))..(((((((((........))))))))-)..))..))))))... ( -30.00) >DroAna_CAF1 8793 87 - 1 --UAUCCCAGCGAGAGC-----------CGGGAAACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG-CCAGCCGGCUC----- --..((((.((....))-----------.))))..((.((((......(((((....)))))..(((((((((........))))))))-))))).))...----- ( -30.80) >consensus __UAUCCCAGCGAGAGCAGAGC______AGGGAAACCUGCUGCAUCAUUUUUCACUCGAAGAUUGCAUAAUGAUUGACAAUUCAUUAUG_CCAGCCAGCUCUUUGC ...........((((((...........(((....)))((((...((.(((((....))))).))((((((((........))))))))..))))..))))))... (-19.57 = -20.90 + 1.33)


| Location | 14,627,480 – 14,627,583 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -25.05 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.03 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14627480 103 - 22407834 UCCGUCCGAGCGUCCGACUGUCUCAGAGUGUGUCUAAAUGUUGGCGGCCCACGCAUUCGCCGGCUAAGAGAUAUCCCAGCGAGAGCAGAGCAGGGAAACCUGC ..((..(....)..)).(((((((...(..(((((....(((((((((....))...)))))))....)))))..)....)))).))).(((((....))))) ( -35.70) >DroEre_CAF1 9527 82 - 1 --------------------UCUCAGAGUCUGUCUAAAUGUUGGCGGCCCACGCAUUUGCCGGCUAAAAGAUAUCCCAGCGAGAGCAGAGCAGGU-AACCUGC --------------------..((.(.(..(((((....((((((.((....))....))))))....)))))..)).((....)).))(((((.-..))))) ( -22.40) >DroYak_CAF1 10352 86 - 1 CCA----------------GUCUCAAAGUCUGUCUAAAUGUUGGCGGCCCACGCAUUCUCCGGCUUAAAGAUAUCCCAGCGAGAGCAGAGCAGGG-AACCUGC ...----------------((((..(((((.(....((((((((....))).)))))..).)))))..))))......((....))...(((((.-..))))) ( -20.90) >consensus _C_________________GUCUCAGAGUCUGUCUAAAUGUUGGCGGCCCACGCAUUCGCCGGCUAAAAGAUAUCCCAGCGAGAGCAGAGCAGGG_AACCUGC ....................(((....(..(((((....(((((((((....))...)))))))....)))))..)..((....)))))(((((....))))) (-25.05 = -25.17 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:38 2006