| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,624,155 – 14,624,246 |
| Length | 91 |
| Max. P | 0.731055 |

| Location | 14,624,155 – 14,624,246 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.34 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14624155 91 + 22407834 AAGUGUAAGCACUGCUAUUAGUUAGAUGAAUAGUAAGGGGAGUGGCACAUGCGGCAUGCAGCAGUAUGUGCAGCUGACUAAGAACUAAAA--------------G--- .((((....))))((((((.........))))))..((..(((.((((((((.((.....)).)))))))).)))..))...........--------------.--- ( -25.90) >DroSec_CAF1 6390 89 + 1 AAGUGC--GCACUGCGAUUAGUUAGAUGAAUAGUAAGGGGAGUGGCACAUGCGGCAAGCAGAAUUAUGUGCAGCUGACUAAAAACUAAAA--------------G--- ..((((--.((((.(..(((.(((......))))))..).))))))))...((((..(((......)))...))))..............--------------.--- ( -20.00) >DroSim_CAF1 6400 89 + 1 AAGUGC--GCACUGCGAUUAGUUAGAUGAAUAGUAAGGGGAGUGGCACAUGCGGCAAGCAGAAUUAUGUGCAGCUGACUAAAAACUAAAA--------------G--- ..((((--.((((.(..(((.(((......))))))..).))))))))...((((..(((......)))...))))..............--------------.--- ( -20.00) >DroEre_CAF1 6439 103 + 1 AAGUGUAAGCACUGCGAUUAGUUAGAUGAAUAGCAAGGGGAGUGGCACAUGCGGCACGGAGA-GUAUGUGCAGCUGACUAAGAACUGAAA-CUUUGUGGAAGUCG--- .((((....)))).(((((.((((......))))..((..(((.((((((((..........-)))))))).)))..))...........-.........)))))--- ( -25.10) >DroYak_CAF1 6533 106 + 1 AAGUGUAAGCACUGCGAUUAGUUAGAUGAAUAGUAAGGGGAGUGGCGAGUGCAUCAUGGAGA-GUAUGUGCAGCUGACUAAGAACUAAAA-GUUUGUGGAAGUCGCAG .((((....))))((((((...(((((...((((..((..(((.(((.((((.((.....))-)))).))).)))..))....))))...-)))))....)))))).. ( -25.90) >DroAna_CAF1 5914 95 + 1 A-GUGC--CCACUGCGGUCGCGGAGAGUCAAGGCGAGGGGACAGGCACU-CUGGAAAAU----GGAGGAGCAGCUGAGCA-GAACUGGCAGAUUUGUGGCGCUC---- (-((((--((.((((....)))).(((((..(((..(....)..((.((-(((.....)----))))..)).)))..((.-......)).)))))).)))))).---- ( -29.00) >consensus AAGUGC__GCACUGCGAUUAGUUAGAUGAAUAGUAAGGGGAGUGGCACAUGCGGCAAGCAGA_GUAUGUGCAGCUGACUAAGAACUAAAA______________G___ .((((....))))....((((((((...................((((((((...........))))))))..))))))))........................... (-14.58 = -14.83 + 0.26)


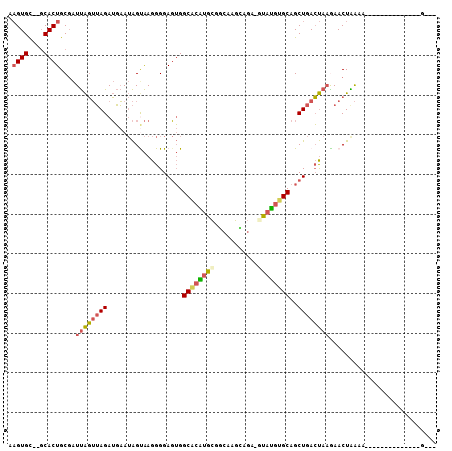
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:34 2006