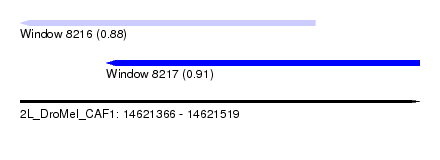
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,621,366 – 14,621,519 |
| Length | 153 |
| Max. P | 0.912970 |

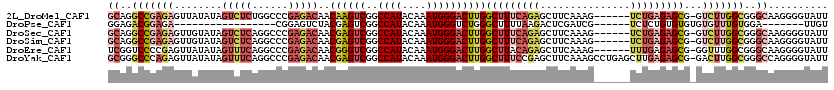
| Location | 14,621,366 – 14,621,479 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -23.33 |
| Energy contribution | -25.89 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14621366 113 - 22407834 GCAGGCCGAGAGUUAUAUAGUCUCUGGCCCGAGACAACAAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAAG------UCUGAGAGCG-GUCUUGGCGGGCAAGGGGUAUU ((.((((((((.(.....).)))).))))((((((..((((((..((((....))))))))))(((((((((.((....))------))))))))).-)))))))).............. ( -45.80) >DroPse_CAF1 4425 90 - 1 GGAGACGGAGA-----------------CGGAGUCUACGAGUCGGCCAUACAAAUGGGUCUGGGCUUUUAAGACUCGAUCG------UCUCUGUGUGUGUGUUUGUGGA-------UUGU ..(.(((((((-----------------((((((((..(((((((((.........))))..)))))...))))))...))------))))))).).............-------.... ( -29.40) >DroSec_CAF1 3557 113 - 1 GCAGGCCGAGAGUUGUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAAG------UCUGAGAGCG-GUCUUGGCGGGCAAGGGGUAUU ((.((((((((.(.....).)))).))))((((((..((((((..((((....))))))))))(((((((((.((....))------))))))))).-)))))))).............. ( -45.50) >DroSim_CAF1 3568 113 - 1 GCAGGCCGAGAGUUGUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAAG------UCUGAGAGCG-GUCUUGGCGGGCAAGGGGUAUU ((.((((((((.(.....).)))).))))((((((..((((((..((((....))))))))))(((((((((.((....))------))))))))).-)))))))).............. ( -45.50) >DroEre_CAF1 3637 113 - 1 UCGGUCCCCGAGUUAUAUAGUUUCAGGCCCGAGACAACGGGUCGGCCAUACAAAUGGGACUUGGCUUACAGAGCUUCAAAG------UUUGAGAGCG-GGUUUGGCGGGCAAGGGGUAUU .....(((((((.........)))..(((((.(((..((((((..((((....))))))))))((((...(((((....))------)))..)))).-.)))...)))))..)))).... ( -36.10) >DroYak_CAF1 3727 119 - 1 GCGGGCCCAGAGUUAUAUAGUUUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCCGAGCUUCAAAGCCUGAGCUUGAGAGCG-GACUUGGCGGGCCAGGGGUAUU ....((((.(((.........))).((((((...(((((((((..((((....))))))))))((((((.((((((........)))))))))))).-...))).))))))..))))... ( -45.50) >consensus GCAGGCCGAGAGUUAUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAAG______UCUGAGAGCG_GUCUUGGCGGGCAAGGGGUAUU ((..(((((((........(((((......)))))..((((((..((((....))))))))))(((((((((...............)))))))))...)))))))..)).......... (-23.33 = -25.89 + 2.56)

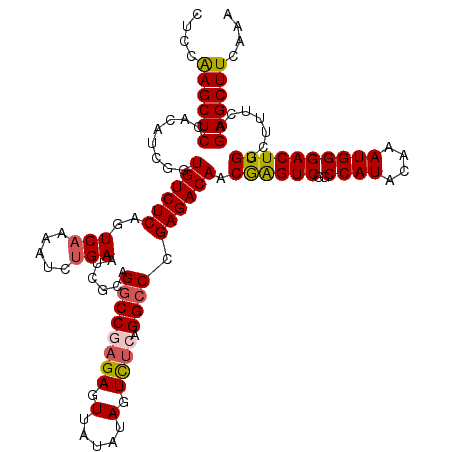
| Location | 14,621,399 – 14,621,519 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -37.52 |
| Consensus MFE | -32.80 |
| Energy contribution | -33.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14621399 120 - 22407834 CUCCAAGCUCCACAUCGGUGUCUCAGUCAAAAUCUGAAUCGCAGGCCGAGAGUUAUAUAGUCUCUGGCCCGAGACAACAAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAA ....((((((....((((((..((((.......))))..))).((((((((.(.....).)))).)))))))((...((((((..((((....))))))))))....)).)))))).... ( -38.80) >DroSec_CAF1 3590 120 - 1 CUCCAAGCUCCACAUCGGUGUCUCAGUCAAAAUCUGAAUCGCAGGCCGAGAGUUGUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAA ....((((((....((((((..((((.......))))..))).((((((((.(.....).)))).)))))))((...((((((..((((....))))))))))....)).)))))).... ( -38.50) >DroSim_CAF1 3601 120 - 1 CUCCAAGCUCCACAUCGGUGUCUCAGUCAAAAUCUGAAUCGCAGGCCGAGAGUUGUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAA ....((((((....((((((..((((.......))))..))).((((((((.(.....).)))).)))))))((...((((((..((((....))))))))))....)).)))))).... ( -38.50) >DroEre_CAF1 3670 120 - 1 CUCCGAGCUCCGCAUCGGUGUCUCAGUCAAAAUCGGAAUCUCGGUCCCCGAGUUAUAUAGUUUCAGGCCCGAGACAACGGGUCGGCCAUACAAAUGGGACUUGGCUUACAGAGCUUCAAA ....((((.(((...))).).)))...........(((.(((.((..(((((((...........((((((......))))))..((((....)))))))))))...)).))).)))... ( -34.70) >DroYak_CAF1 3766 120 - 1 CUCCAAGCUCCGCAUCGGUGUCUCAGUCAAAAUCUGAAUCGCGGGCCCAGAGUUAUAUAGUUUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCCGAGCUUCAAA ...........((.((((((((((..(((.....))).....(((((..(((.........))).))))))))))).((((((..((((....)))))))))).....))))))...... ( -37.10) >consensus CUCCAAGCUCCACAUCGGUGUCUCAGUCAAAAUCUGAAUCGCAGGCCGAGAGUUAUAUAGUCUCAGGCCCGAGACAACGAGUCGGCCAUACAAAUGGGACUUGGCUUUCAGAGCUUCAAA ....((((((........((((((..(((.....)))......((((((((.(.....).)))).)))).)))))).((((((..((((....)))))))))).......)))))).... (-32.80 = -33.08 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:31 2006