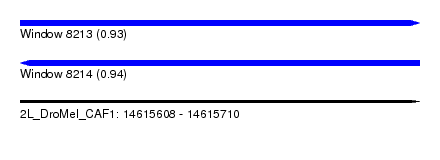
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,615,608 – 14,615,710 |
| Length | 102 |
| Max. P | 0.941892 |

| Location | 14,615,608 – 14,615,710 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14615608 102 + 22407834 UACUUGCAUCUCUUCAAAUUUACUUAAUUGAUCAAGUAAGUAGCAAAAGGGCACACAAUUGAAGGAAAUUCUUGUUUAAUUGAAUUUAUUAUGCAAGUGCGG (((((((((.(((((((((((((((........)))))))).((......))......)))))))((((((..........))))))...)))))))))... ( -24.60) >DroSec_CAF1 24038 102 + 1 UACUUGCAUCUCUUCAAAUUUACUUAAUUGAUCAAGUAAGUAGCAAAAGGGCGACCAAUUGAAGGAAAUUCUUGUUUAAUUUAAUUUAUAAUGCAAGUGCGG (((((((((.(((((((((((((((........)))))))).......((....))..)))))))((((....)))).............)))))))))... ( -23.60) >DroSim_CAF1 29316 102 + 1 UACUUGCAUCUCUUCAAAUUUACUUAAUUGAUCAAGUAAGUAGCAAGAGGGCGACCAAUUGAAGGAAAUUCUUGUUUAAAUGAAUUUAUUAUGCAAGUGCGG (((((((((.(((((((((((((((........)))))))).......((....))..)))))))((((((..........))))))...)))))))))... ( -26.80) >consensus UACUUGCAUCUCUUCAAAUUUACUUAAUUGAUCAAGUAAGUAGCAAAAGGGCGACCAAUUGAAGGAAAUUCUUGUUUAAUUGAAUUUAUUAUGCAAGUGCGG (((((((((.(((((((((((((((........)))))))).((......))......)))))))((((((..........))))))...)))))))))... (-22.83 = -23.17 + 0.33)

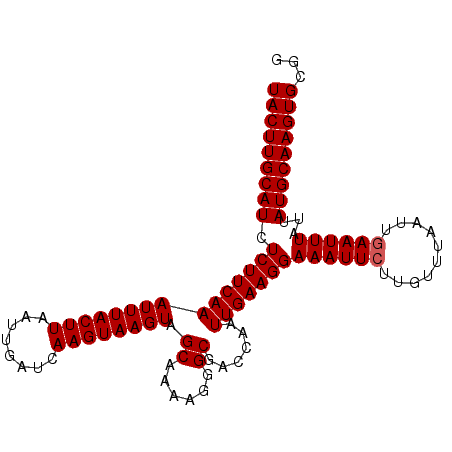
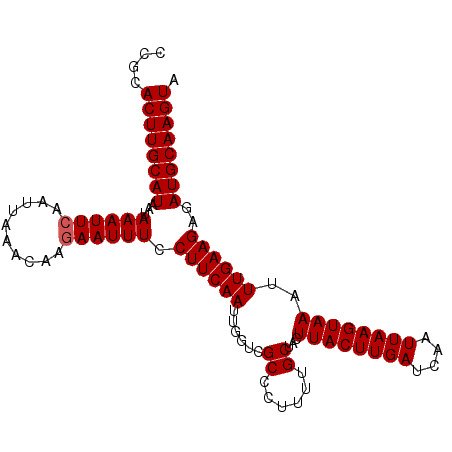
| Location | 14,615,608 – 14,615,710 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -22.43 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14615608 102 - 22407834 CCGCACUUGCAUAAUAAAUUCAAUUAAACAAGAAUUUCCUUCAAUUGUGUGCCCUUUUGCUACUUACUUGAUCAAUUAAGUAAAUUUGAAGAGAUGCAAGUA ....((((((((...((((((..........)))))).((((((..(((.((......)))))((((((((....))))))))..))))))..)))))))). ( -23.20) >DroSec_CAF1 24038 102 - 1 CCGCACUUGCAUUAUAAAUUAAAUUAAACAAGAAUUUCCUUCAAUUGGUCGCCCUUUUGCUACUUACUUGAUCAAUUAAGUAAAUUUGAAGAGAUGCAAGUA ....(((((((((.......(((((.......))))).((((((...((.((......)).))((((((((....))))))))..)))))).))))))))). ( -21.30) >DroSim_CAF1 29316 102 - 1 CCGCACUUGCAUAAUAAAUUCAUUUAAACAAGAAUUUCCUUCAAUUGGUCGCCCUCUUGCUACUUACUUGAUCAAUUAAGUAAAUUUGAAGAGAUGCAAGUA ....((((((((...((((((..........)))))).((((((...((.((......)).))((((((((....))))))))..))))))..)))))))). ( -22.80) >consensus CCGCACUUGCAUAAUAAAUUCAAUUAAACAAGAAUUUCCUUCAAUUGGUCGCCCUUUUGCUACUUACUUGAUCAAUUAAGUAAAUUUGAAGAGAUGCAAGUA ....((((((((...((((((..........)))))).((((((......((......))...((((((((....))))))))..))))))..)))))))). (-20.07 = -20.40 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:28 2006