| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,602,757 – 14,602,877 |
| Length | 120 |
| Max. P | 0.962600 |

| Location | 14,602,757 – 14,602,877 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -25.24 |
| Energy contribution | -25.85 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
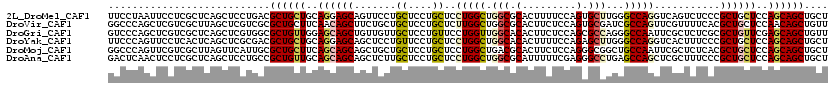
>2L_DroMel_CAF1 14602757 120 + 22407834 UUCCUAAUUCCUCGCUCAGCUCCUGACGCUGCUGCAGGAGCAGUUCCUGCUCCUGCUCCUGGCUGGCGCACUUUUCCAGUGCUUGGGCCAGGUCAGUCUCCCGCUGCUCCAGCAGCUGCU ................(((((.(((..((.((.((((((((((...)))))))))).(((((((.(.(((((.....))))).).)))))))..........)).))..))).))))).. ( -54.60) >DroVir_CAF1 1428 120 + 1 GGCCCAGCUCGUCGCUUAGCUCGUCGCGCUGCUUCAACAGCUUCUGCUGCUCCUGCUCUUGGCUGGCGCACUUCUCCAGUGCGAUCGCCAGUUCGUUUUCACGCUGCUCCAACAGCUGUU ((..((((.((.((.......)).)).))))..))(((((((..((..((....((..(((((...((((((.....))))))...)))))...((....)))).))..))..))))))) ( -37.00) >DroGri_CAF1 7588 120 + 1 GUCCCAGCUCGUCGCUCAGCUCGUGGCGCUGUUGGAGCAGCUGUUGUUGCUCCUGUUCCUGGUUGGCACACUUCUCCAGCGCCAGGGCCAAUUCGCUCUCGCGCUGUUCGAGCAGCUGUU ....(((((..(((..((((.((((((((((..((((((((....))))))))(((.((.....)).)))......))))))))((((......)))).)).))))..)))..))))).. ( -55.00) >DroYak_CAF1 11345 120 + 1 UUCCCAGUUCCUCACUCAGCUCGCGACGCUGCUGCAGGAGCAGCUCCUGUUCCUGCUCCUGGCUGGCACACUUUUCCAGAGCUUGGGCCAGGUCACUUUCCCGCUGCUCCAGCAGCUGCU .(((.((((........)))).).)).((.(((((.((((((((.((((..((.((((.(((..((....))...)))))))..))..))))..........)))))))).))))).)). ( -47.00) >DroMoj_CAF1 7652 120 + 1 GGCCCAGUUCGUCGCUUAGUUCAUUGCGCUGCUUCAGCAGCAGCUGCUGCUCCUGCUCCUGGCUGACGCACUUCUCCAGGGCGGCUGCCAAUUCGCUCUCACGCUGCUCCAGCAGCUGCU (((.......)))............((((((((..((((((((((((.((....)).(((((..((......)).)))))))))))((......))......))))))..)))))).)). ( -43.20) >DroAna_CAF1 1971 120 + 1 GACUCAACUCCUCGCUCAGCUCCUGCCGCUGUUGCAGCAGCAGCUCUUGCUCCUGCUCCUGGCUGGCGCAUUUUUCGAGGGCCUGAGCCAGCUCGCUUUCCCGCUGCUCCAGCAGCUGCU (.((((..((((((....((.((.(((((....))(((((.(((....))).)))))...))).)).))......))))))..)))).).............(((((....))))).... ( -44.70) >consensus GUCCCAGCUCCUCGCUCAGCUCGUGGCGCUGCUGCAGCAGCAGCUCCUGCUCCUGCUCCUGGCUGGCGCACUUCUCCAGGGCCUGGGCCAGUUCGCUCUCCCGCUGCUCCAGCAGCUGCU ...........................((((((..((((((.......((....))..(((((.(((.(.........).)))...)))))...........))))))..)))))).... (-25.24 = -25.85 + 0.61)

| Location | 14,602,757 – 14,602,877 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -51.37 |
| Consensus MFE | -28.04 |
| Energy contribution | -28.43 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14602757 120 - 22407834 AGCAGCUGCUGGAGCAGCGGGAGACUGACCUGGCCCAAGCACUGGAAAAGUGCGCCAGCCAGGAGCAGGAGCAGGAACUGCUCCUGCAGCAGCGUCAGGAGCUGAGCGAGGAAUUAGGAA ..(((((.(((..((.((((....))..((((((....(((((.....)))))....)))))).((((((((((...)))))))))).)).))..))).)))))................ ( -61.90) >DroVir_CAF1 1428 120 - 1 AACAGCUGUUGGAGCAGCGUGAAAACGAACUGGCGAUCGCACUGGAGAAGUGCGCCAGCCAAGAGCAGGAGCAGCAGAAGCUGUUGAAGCAGCGCGACGAGCUAAGCGACGAGCUGGGCC ..(((((((((.(((.(((((.........((((...((((((.....))))))...))))...((...((((((....))))))...))..)))).)..)))...)))).))))).... ( -45.00) >DroGri_CAF1 7588 120 - 1 AACAGCUGCUCGAACAGCGCGAGAGCGAAUUGGCCCUGGCGCUGGAGAAGUGUGCCAACCAGGAACAGGAGCAACAACAGCUGCUCCAACAGCGCCACGAGCUGAGCGACGAGCUGGGAC ..(((((..(((..((((((....))........(.((((((((......(((.((.....)).)))((((((........))))))..)))))))).).))))..)))..))))).... ( -52.00) >DroYak_CAF1 11345 120 - 1 AGCAGCUGCUGGAGCAGCGGGAAAGUGACCUGGCCCAAGCUCUGGAAAAGUGUGCCAGCCAGGAGCAGGAACAGGAGCUGCUCCUGCAGCAGCGUCGCGAGCUGAGUGAGGAACUGGGAA .((.(((((.(((((((((((..((....))..)))..(((((((....(....)...))).))))..........)))))))).))))).))........((.(((.....))).)).. ( -49.70) >DroMoj_CAF1 7652 120 - 1 AGCAGCUGCUGGAGCAGCGUGAGAGCGAAUUGGCAGCCGCCCUGGAGAAGUGCGUCAGCCAGGAGCAGGAGCAGCAGCUGCUGCUGAAGCAGCGCAAUGAACUAAGCGACGAACUGGGCC (((((((((((..((..(((....))).....))..(((((((((.((......))..))))).)).))..)))))))))))(((..((..((((..........))).)...)).))). ( -45.80) >DroAna_CAF1 1971 120 - 1 AGCAGCUGCUGGAGCAGCGGGAAAGCGAGCUGGCUCAGGCCCUCGAAAAAUGCGCCAGCCAGGAGCAGGAGCAAGAGCUGCUGCUGCAACAGCGGCAGGAGCUGAGCGAGGAGUUGAGUC (((.(((((....)))))(......)..)))(((((((..(((((.....(((.((.((.....)).)).)))..((((.(((((((....))))))).))))...)))))..))))))) ( -53.80) >consensus AGCAGCUGCUGGAGCAGCGGGAAAGCGAACUGGCCCAAGCACUGGAAAAGUGCGCCAGCCAGGAGCAGGAGCAGCAGCUGCUGCUGCAGCAGCGCCACGAGCUGAGCGACGAACUGGGAC ....((((((..((((((((....))...(((((....((.((.....)).))....))))).................))))))..))))))........((.(((.....))).)).. (-28.04 = -28.43 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:21 2006