| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,602,244 – 14,602,348 |
| Length | 104 |
| Max. P | 0.969941 |

| Location | 14,602,244 – 14,602,348 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.73 |
| Mean single sequence MFE | -24.55 |
| Consensus MFE | -14.92 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
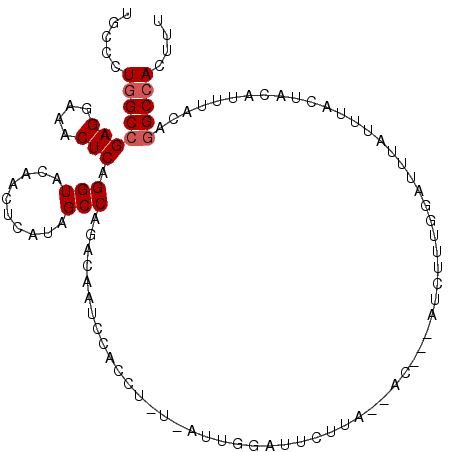
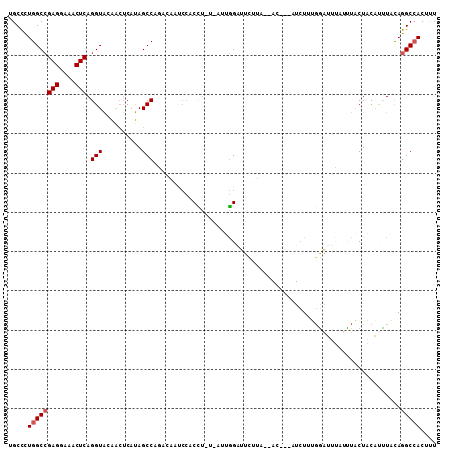
>2L_DroMel_CAF1 14602244 104 - 22407834 UGCCCUGGCCGAGGAAACUCAGGUACAGCUCAUAGCCCGCCAAUCCACCUUUGUUUGGAUUCUUA--AC---UUCUUUGGAUUUAUUUACUACAUUUACAGGCCACUUU .....((((((((....))).(((...((.....))..)))((((((.....(((.((...)).)--))---.....)))))).................))))).... ( -24.60) >DroPse_CAF1 14817 99 - 1 CGCUUUGGCAGAGGAAACUCAGGUGGGAAACACAGCCACA-------CU--U-UUGAGAGUCCAAGUACCUAAUCUUGGAUUGGUUUUCCUCCAUUUCCAGGCAACCCU .((((.((..(((....))).((.(((((((.(((.....-------..--.-)))..((((((((........)))))))).))))))).))....))))))...... ( -32.70) >DroSec_CAF1 10723 103 - 1 UGCCCUGGCCGAGGAAACUCAGGUACAGCUCAUAGCCCACCAAUCCACCUUU-AUUGGAUUCUUA--AC---UUCUUUGGAUUUAUUUACUACAUUUACAGGCCACUUU .....((((((((....))).(((...((.....))..)))((((((.....-..))))))....--..---............................))))).... ( -23.30) >DroSim_CAF1 11014 103 - 1 UGCCCUGGCCGAGGAAACUCAGGUACAGCUCAUAGCCCACCAAUCCACCUUU-UUUGGAUUUUUA--AC---UUCUUUGGAUUUAUUUACUACAUUUACAGGCCACUUU .....((((((((....))).(((...((.....))..)))((((((.....-..))))))....--..---............................))))).... ( -23.30) >DroEre_CAF1 8562 101 - 1 CGCCCUGGCCGAGGAAACUCAGGUAAAACUCAUAGCCAGACAACCCCCCU-C-A-UGGUUUCUUA--AC---AUCUUUGGACACUUCUUCUUCGCCUUUAGGCCACGUU .....((((((((....)))((((.............(((.((((.....-.-.-.)))))))..--.(---(....))..............))))...))))).... ( -22.00) >DroYak_CAF1 10869 102 - 1 UGCCCUGGCCGAGGAAACUCAGGUAAAACUCAUAGCCAGACAGCCCCCUU-U-AGUGGUUUCUUU--AC---AUAUUUCGAUACUUUUACUGCAUUUAUAGGCCACUUU .....((((((((....))).(((((((.((...((......))......-.-.((((.....))--))---.......))...))))))).........))))).... ( -21.40) >consensus UGCCCUGGCCGAGGAAACUCAGGUACAACUCAUAGCCAGACAAUCCACCU_U_AUUGGAUUCUUA__AC___AUCUUUGGAUUUAUUUACUACAUUUACAGGCCACUUU .....((((((((....))).(((..........)))...............................................................))))).... (-14.92 = -15.25 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:19 2006