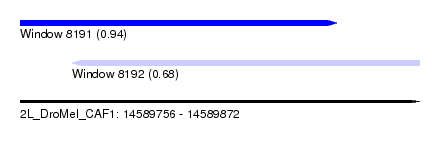
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,589,756 – 14,589,872 |
| Length | 116 |
| Max. P | 0.943247 |

| Location | 14,589,756 – 14,589,848 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14589756 92 + 22407834 UUGUCAGCC-AGAUCU------------------------CCACACAAACCAAACCGAUCCU-AUUCGAAUUGAAACCCGCCGCAUCUAUCAGCGGCGACAGA-AACACUUUGC-CUGUC ....(((.(-(((...------------------------..........(((..(((....-..)))..))).....((((((........)))))).....-.....)))).-))).. ( -19.40) >DroSec_CAF1 132001 92 + 1 UUGUCAGCC-AGAUCU------------------------CCACACAAACCAAACCGAUCCU-AUUCGAAUCGAAACCCGCCGCAUCUAUCAGCGGCGACAGA-AACACUUUGC-CUGUC ....(((.(-(((...------------------------...............((((.(.-....).)))).....((((((........)))))).....-.....)))).-))).. ( -18.30) >DroSim_CAF1 110296 93 + 1 UUGUCAGCCGAGAUCU------------------------CCACACAAACCAAACCGAUCCU-AUUCGAAUCGAAACCCGCCGCAUCUAUCAGCGGCGACAGA-AACACUUUGC-CUGUC ....((((.(((....------------------------...............((((.(.-....).)))).....((((((........)))))).....-....))).).-))).. ( -18.00) >DroEre_CAF1 116576 96 + 1 UUGUCAGCC-AGCUCUCCAU--------------------CCACACAAGCAGAAUUGAUCCC-AUUCGAAUCGGAAC-CGCCGCAUCUAUCAGCGGCGACAGA-AACACUUUGGCUUGUU .........-..........--------------------....((((((.((.((((....-..)))).)).....-((((((........)))))).((((-.....)))))))))). ( -23.60) >DroYak_CAF1 115330 116 + 1 UUGUCAGCC-AGAUCUGCAUCUCCAUCUCCAUCUCCAUUUCCACACAAACCAAACUGAUGCU-AUUGGAAUCGAAACCCGCCGCAUCUAUCAGCGGCGACAGA-AACACUUUGC-CUGUC ....(((.(-(((((((....((((....((((.......................))))..-..)))).........((((((........)))))).))))-.....)))).-))).. ( -24.70) >DroAna_CAF1 57531 84 + 1 UUGUCAGGC-AGAGGA---------------------------------CC-AACAGGUCCGGAUCGGAAUCGAAACCAGCCGCAUCGAUCAGCGGCGACAGAAAACACUUUGC-CUGCU ..(.(((((-((((((---------------------------------((-....)))))((.(((....)))..)).(((((........)))))............)))))-)))). ( -32.90) >consensus UUGUCAGCC_AGAUCU________________________CCACACAAACCAAACCGAUCCU_AUUCGAAUCGAAACCCGCCGCAUCUAUCAGCGGCGACAGA_AACACUUUGC_CUGUC (((((..................................................((((.(......).))))......(((((........)))))))))).................. (-16.06 = -15.78 + -0.28)

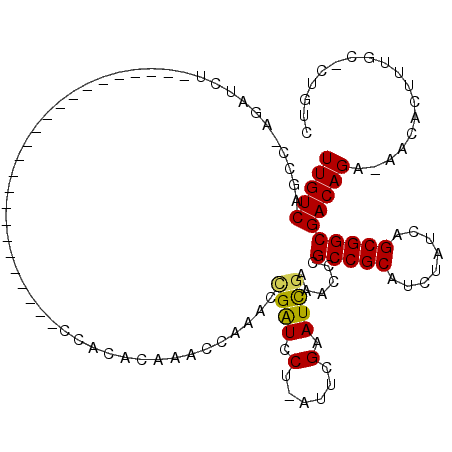
| Location | 14,589,771 – 14,589,872 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 67.43 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -23.23 |
| Energy contribution | -25.10 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14589771 101 - 22407834 CAA--------------GAAACCAACAAGCCGGCAGACGACAG-GCAAAGUGUU-UCUGUCGCCGCUGAUAGAUGCGGCGGGUUUCAAUUCGAAU-AGGAUCGGUUUGGUUUGUGUGG ...--------------....((((((((((((((((.((((.-......))))-))))))(((((........)))))..........((((..-....))))...))))))).))) ( -33.00) >DroSim_CAF1 110312 101 - 1 CAA--------------GAAACCAACAAGCCGGCAGACGACAG-GCAAAGUGUU-UCUGUCGCCGCUGAUAGAUGCGGCGGGUUUCGAUUCGAAU-AGGAUCGGUUUGGUUUGUGUGG ...--------------....((((((((((((((((.((((.-......))))-))))))(((((........))))).....(((((((....-.)))))))...))))))).))) ( -38.60) >DroEre_CAF1 116595 104 - 1 CAAGCCAGCCAAGCCAGCCA-----------AGCAUCCAACAAGCCAAAGUGUU-UCUGUCGCCGCUGAUAGAUGCGGCG-GUUCCGAUUCGAAU-GGGAUCAAUUCUGCUUGUGUGG ....(((..(((((..(((.-----------.(((((.....(((....(((..-.....))).)))....))))))))(-((((((.......)-))))))......)))))..))) ( -29.80) >DroAna_CAF1 57546 93 - 1 CCC--------------AGACCCAAACGAUCGACGGACAGCAG-GCAAAGUGUUUUCUGUCGCCGCUGAUCGAUGCGGCUGGUUUCGAUUCCGAUCCGGACCUGUU-GG--------- ...--------------....((((.((((((.(((((((.((-((.....)))).))))..))).))))))....((((((..(((....))).)))).))..))-))--------- ( -30.10) >consensus CAA______________GAAACCAACAAGCCGGCAGACAACAG_GCAAAGUGUU_UCUGUCGCCGCUGAUAGAUGCGGCGGGUUUCGAUUCGAAU_AGGAUCGGUUUGGUUUGUGUGG .................((((((........((((((.((((........)))).))))))(((((........))))).))))))(((((......)))))................ (-23.23 = -25.10 + 1.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:07 2006