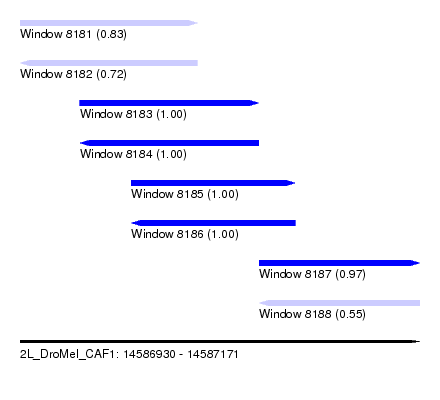
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,586,930 – 14,587,171 |
| Length | 241 |
| Max. P | 0.999817 |

| Location | 14,586,930 – 14,587,037 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.42 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
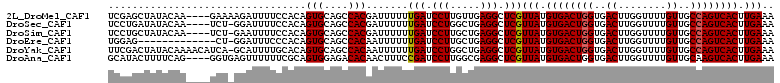
>2L_DroMel_CAF1 14586930 107 + 22407834 UCGAGCUAUACAA----GAAAAGAUUUUCCACAGUGCAGCCACGAUUUUUUGAUCCUUGUUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA .((((((..((((----(...............(((....)))((((....)))))))))...))))))(((.((((((((..((........))..)))))))).))).. ( -37.20) >DroSec_CAF1 128656 106 + 1 UCCUGAUAUACAA----UCU-GGAUUUUCCACAGUGCAGCCACGAUUUUUUGAUCCUGGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA (((.(((.....)----)).-))).((((.(((((.((((((.((((....)))).))))))..))).))...((((((((..((........))..))))))))..)))) ( -39.30) >DroSim_CAF1 106423 106 + 1 UCCUGCUAUACAA----UCU-GAAUUUUCCACAGUGCAGCCACGAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA ....(((......----.((-(.........)))..((((...((((....))))...)))).)))...(((.((((((((..((........))..)))))))).))).. ( -29.70) >DroEre_CAF1 112288 97 + 1 UGGAG-------------CU-GGAUUUCCCACAGUGCAGCCACAAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA ..(((-------------((-((.....)).(((((.((...(((....)))...))))))).))))).(((.((((((((..((........))..)))))))).))).. ( -33.20) >DroYak_CAF1 110835 110 + 1 UUCGACUAUACAAAACAUCA-GCAUUUUGCACAGUGCAGCCACAAUUUUUUGAUCCUGGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA (((((........(((....-((.....))..(((.((((((..(((....)))..))))))..))).)))..((((((((..((........))..))))))))))))). ( -35.90) >DroAna_CAF1 55677 107 + 1 GCAUACUUUUCAG----GGUGAGUUUUUUCGCAGUGGAGACACAACUUUCCGAUCCUUGGCGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCAAGUCACUUGAAA .....(((.((((----((((((....))))).(((....)))............))))).))).....(((.((((((.(..((........))..).)))))).))).. ( -31.00) >consensus UCCUGCUAUACAA____UCU_GGAUUUUCCACAGUGCAGCCACAAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAA .................................(((....))).......(((.(((.....))).)))(((.((((((((..((........))..)))))))).))).. (-25.39 = -25.42 + 0.03)
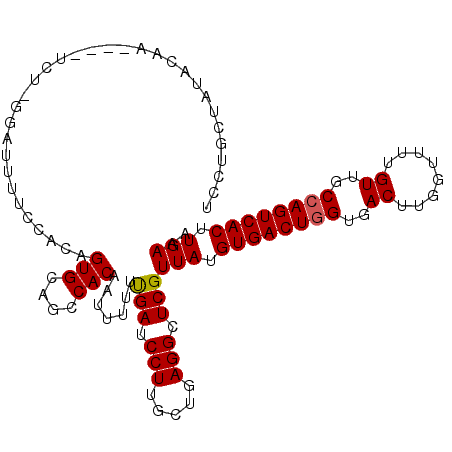
| Location | 14,586,930 – 14,587,037 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.00 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14586930 107 - 22407834 UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAACAAGGAUCAAAAAAUCGUGGCUGCACUGUGGAAAAUCUUUUC----UUGUAUAGCUCGA ......((((((((..((........))..))))))))....(((((....(((((((........(((..(.....)..)))........)))----))))...))))). ( -31.39) >DroSec_CAF1 128656 106 - 1 UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCCAGGAUCAAAAAAUCGUGGCUGCACUGUGGAAAAUCC-AGA----UUGUAUAUCAGGA ......((((((((..((........))..))))))))........(((.(((((.(((......))).)))))(((.(.(((.....))-).)----.))).....))). ( -33.30) >DroSim_CAF1 106423 106 - 1 UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUCGUGGCUGCACUGUGGAAAAUUC-AGA----UUGUAUAGCAGGA ......((((((((..((........))..))))))))........(((((((.(.(((......))).).))))..(((((.((.....-...----)).))))).))). ( -25.70) >DroEre_CAF1 112288 97 - 1 UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUUGUGGCUGCACUGUGGGAAAUCC-AG-------------CUCCA ......((((((((..((........))..)))))))).....((((..((((.(.(((......))).).)))).......((....))-.)-------------))).. ( -26.50) >DroYak_CAF1 110835 110 - 1 UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCCAGGAUCAAAAAAUUGUGGCUGCACUGUGCAAAAUGC-UGAUGUUUUGUAUAGUCGAA ......((((((((..((........))..))))))))....(((....((((((.(((......))).))))))..(((((((((((..-....)))))))))))))).. ( -39.00) >DroAna_CAF1 55677 107 - 1 UUUCAAGUGACUUGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCGCCAAGGAUCGGAAAGUUGUGUCUCCACUGCGAAAAAACUCACC----CUGAAAAGUAUGC ((((..((((((((.........))))))))((((.(((((((...((...(....)...))...)))))))....)))).)))).....((..----((....))..)). ( -24.70) >consensus UUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUCGUGGCUGCACUGUGGAAAAUCC_AGA____UUGUAUAGCAGGA (((((.((((((((..((........))..)))))))).....((.(((.....))).))........(((....)))..))))).......................... (-19.13 = -19.38 + 0.25)


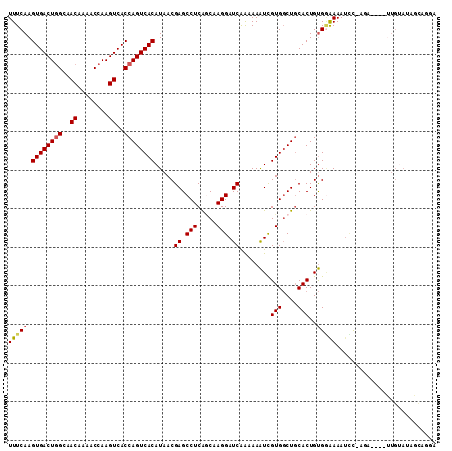
| Location | 14,586,966 – 14,587,074 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -34.44 |
| Consensus MFE | -31.07 |
| Energy contribution | -31.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.68 |
| SVM RNA-class probability | 0.999515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14586966 108 + 22407834 CACGAUUUUUUGAUCCUUGUUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGC ...........((((((((((((((((..(((.((((((((..((........))..)))))))).)))..)))).))))).......))))))).(((.....))). ( -35.61) >DroSec_CAF1 128691 108 + 1 CACGAUUUUUUGAUCCUGGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGC ...(((((((..(...((..(((((((..(((.((((((((..((........))..)))))))).)))..)))).)))..))..)..))))))).(((.....))). ( -36.30) >DroSim_CAF1 106458 108 + 1 CACGAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGC ...........(((((((.....((((..(((.((((((((..((........))..)))))))).)))..))))..(((.....)))))))))).(((.....))). ( -34.10) >DroEre_CAF1 112314 108 + 1 CACAAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUGGGGGGAUUUCGCUUUUUGCGC ...........(((((((.((((((((..(((.((((((((..((........))..)))))))).)))..)))).....)))...).))))))).(((.....))). ( -35.60) >DroYak_CAF1 110874 108 + 1 CACAAUUUUUUGAUCCUGGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUCCGCUUUUUGCGC ...(((((((..(...((..(((((((..(((.((((((((..((........))..)))))))).)))..)))).)))..))..)..))))))).(((.....))). ( -36.10) >DroAna_CAF1 55713 107 + 1 CACAACUUUCCGAUCCUUGGCGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCAAGUCACUUGAAAAGUCAUUAAUCAUUUCG-GGGAUUCCGCUUUUUGCAC ......(..((((..........((((..(((.((((((.(..((........))..).)))))).)))..))))..........)))-)..)....((.....)).. ( -28.95) >consensus CACAAUUUUUUGAUCCUUGCUGAGGCUCGUUAUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGC ...........((((((......((((..(((.((((((((..((........))..)))))))).)))..)))).....((.....)))))))).(((.....))). (-31.07 = -31.57 + 0.50)

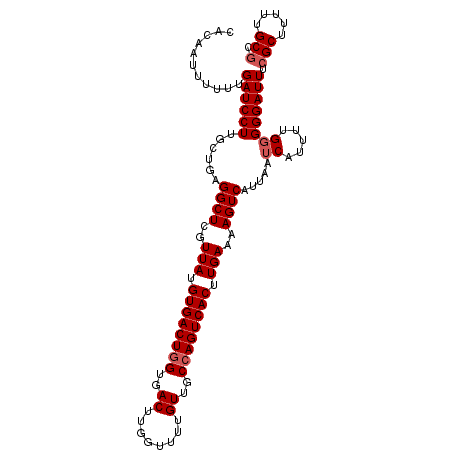
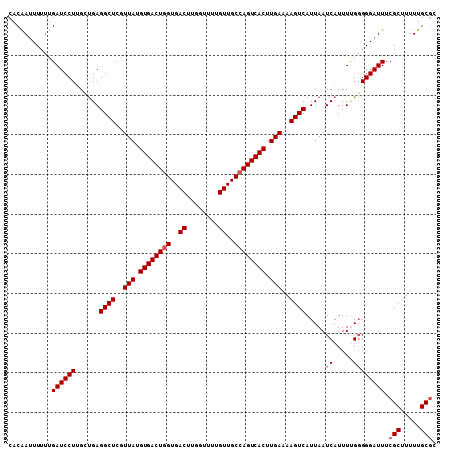
| Location | 14,586,966 – 14,587,074 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -22.66 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14586966 108 - 22407834 GCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAACAAGGAUCAAAAAAUCGUG .(((.....)))............((((((.............((((((((..((........))..)))))))).....((.(((.....))).))....)))))). ( -24.90) >DroSec_CAF1 128691 108 - 1 GCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCCAGGAUCAAAAAAUCGUG .(((.....)))............((((((.............((((((((..((........))..)))))))).....((.(((.....))).))....)))))). ( -24.80) >DroSim_CAF1 106458 108 - 1 GCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUCGUG .(((.....)))............((((((.............((((((((..((........))..)))))))).....((.(((.....))).))....)))))). ( -24.90) >DroEre_CAF1 112314 108 - 1 GCGCAAAAAGCGAAAUCCCCCCAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUUGUG .(((.....))).............(((((.............((((((((..((........))..)))))))).......((....))...))))).......... ( -22.20) >DroYak_CAF1 110874 108 - 1 GCGCAAAAAGCGGAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCCAGGAUCAAAAAAUUGUG .(((((.....((......))....(((((..((((((.....((((((((..((........))..)))))))).....)))..))).....))))).....))))) ( -23.60) >DroAna_CAF1 55713 107 - 1 GUGCAAAAAGCGGAAUCCC-CGAAAUGAUUAAUGACUUUUCAAGUGACUUGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCGCCAAGGAUCGGAAAGUUGUG ..........(((.....)-)).........(..(((((((..((((((((.........))))))))............((.(((.....))).)))))))))..). ( -24.80) >consensus GCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAUAACGAGCCUCAGCAAGGAUCAAAAAAUCGUG .(((.....)))............((((((.............((((((((..((........))..)))))))).....((.(((.....))).))....)))))). (-22.66 = -22.30 + -0.36)

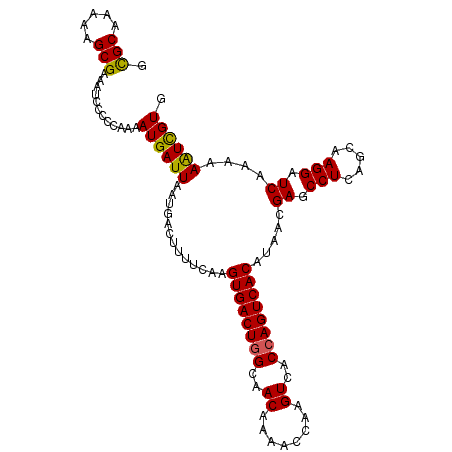
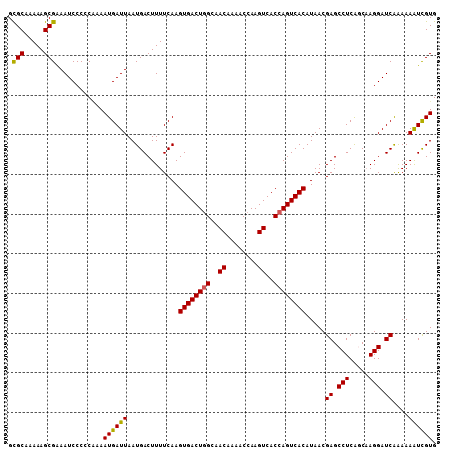
| Location | 14,586,997 – 14,587,096 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.78 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.999187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14586997 99 + 22407834 AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGCCAUUUGGGGCGUAGUUGUGCGC ..((((((((..((........))..)))))))).....(((((.((((.....))))..)))))(((...(((((((......)))))))....))). ( -35.50) >DroSec_CAF1 128722 99 + 1 AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGCCAUUUGGGGCGUAGUUGUGCGC ..((((((((..((........))..)))))))).....(((((.((((.....))))..)))))(((...(((((((......)))))))....))). ( -35.50) >DroSim_CAF1 106489 99 + 1 AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGCCAUUUGGGGCGUAGUUGUGCGC ..((((((((..((........))..)))))))).....(((((.((((.....))))..)))))(((...(((((((......)))))))....))). ( -35.50) >DroEre_CAF1 112345 99 + 1 AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUGGGGGGAUUUCGCUUUUUGCGCCAUUUCGGGCGUAGCUGUGCGC ..((((((((..((........))..)))))))).....(((((.((((....))))...)))))(((...(((((((......)))))))....))). ( -34.80) >DroYak_CAF1 110905 99 + 1 AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUCCGCUUUUUGCGCCAUUUGGGGCGUAGUUGUGCGC ..((((((((..((........))..))))))))......((((.((((.....))))..)))).(((...(((((((......)))))))....))). ( -34.80) >DroAna_CAF1 55744 98 + 1 AUGUGACUGGUGACUUGGUUUUGUUGCAAGUCACUUGAAAAGUCAUUAAUCAUUUCG-GGGAUUCCGCUUUUUGCACCAUUUGGGGCGGAGUUGUGUGC ..(((((((((((((((.........))))))))).....))))))...........-(.(((((((((((...........))))))))))).).... ( -29.20) >consensus AUGUGACUGGUGACUUGGUUUUGUUGCCAGUCACUUGAAAAGUCAUUAAUCAUUUUGGGGGAUUUCGCUUUUUGCGCCAUUUGGGGCGUAGUUGUGCGC ..((((((((..((........))..))))))))...............................(((...(((((((......)))))))....))). (-30.42 = -30.78 + 0.36)


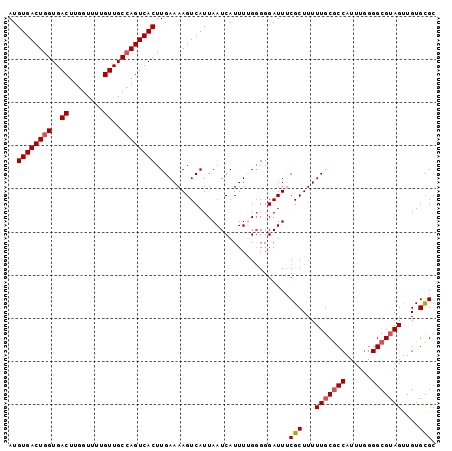
| Location | 14,586,997 – 14,587,096 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14586997 99 - 22407834 GCGCACAACUACGCCCCAAAUGGCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. ( -23.32) >DroSec_CAF1 128722 99 - 1 GCGCACAACUACGCCCCAAAUGGCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. ( -23.32) >DroSim_CAF1 106489 99 - 1 GCGCACAACUACGCCCCAAAUGGCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. ( -23.32) >DroEre_CAF1 112345 99 - 1 GCGCACAGCUACGCCCGAAAUGGCGCAAAAAGCGAAAUCCCCCCAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. ( -23.32) >DroYak_CAF1 110905 99 - 1 GCGCACAACUACGCCCCAAAUGGCGCAAAAAGCGGAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. ( -23.52) >DroAna_CAF1 55744 98 - 1 GCACACAACUCCGCCCCAAAUGGUGCAAAAAGCGGAAUCCC-CGAAAUGAUUAAUGACUUUUCAAGUGACUUGCAACAAAACCAAGUCACCAGUCACAU ((((.((.............))))))......(((.....)-))..........(((((......((((((((.........)))))))).)))))... ( -19.42) >consensus GCGCACAACUACGCCCCAAAUGGCGCAAAAAGCGAAAUCCCCCAAAAUGAUUAAUGACUUUUCAAGUGACUGGCAACAAAACCAAGUCACCAGUCACAU .(((.......((((......))))......)))...............................((((((((..((........))..)))))))).. (-21.01 = -21.20 + 0.20)


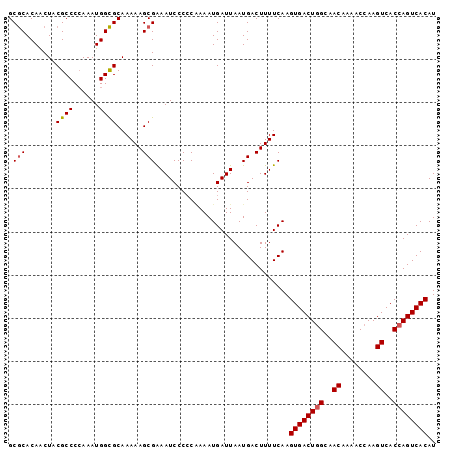
| Location | 14,587,074 – 14,587,171 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.71 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -35.67 |
| Energy contribution | -35.95 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14587074 97 + 22407834 CAUUUGGGGCGUAGUUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGGAGUGCUGCAGAAAUCCAUCACGCACCCAAAUGGCAGCCACUCG---- ((((((((((((.((.(..(.((((...(((((((((....))))))))))))).)..).)).((......)))))).))))))))...........---- ( -40.50) >DroSec_CAF1 128799 97 + 1 CAUUUGGGGCGUAGUUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGGAGUGCUGCAGAAAUCCAUCACGCACCCAAAUGGCAGCCACUCG---- ((((((((((((.((.(..(.((((...(((((((((....))))))))))))).)..).)).((......)))))).))))))))...........---- ( -40.50) >DroSim_CAF1 106566 97 + 1 CAUUUGGGGCGUAGUUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGGAGUGCUGCAGAAAUCCAUCACGCACCCAAAUGGCAGCCACUCG---- ((((((((((((.((.(..(.((((...(((((((((....))))))))))))).)..).)).((......)))))).))))))))...........---- ( -40.50) >DroEre_CAF1 112422 97 + 1 CAUUUCGGGCGUAGCUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGGAGUGCUGCAGAAAUCCAUCACGCGCCCAAAUGGCAGCCACUCG---- (((((.((((((.((.(..(.((((...(((((((((....))))))))))))).)..).)).((......))..)))))))))))...........---- ( -39.10) >DroYak_CAF1 110982 101 + 1 CAUUUGGGGCGUAGUUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGUAGCGCUGCAGAAAUCCAUCACGCACCCAAAUGGCAGCCACUCGCAUC ((((((((((((.((.((((..(((...(((((((((....))))))))))))..)))).)).((......)))))).))))))))((........))... ( -40.30) >DroAna_CAF1 55820 97 + 1 CAUUUGGGGCGGAGUUGUGUGCCGAGCUGUCGGAGUUGCUGGAUUUCGACUCAGAGUGCUGCAGAAAUCCAUCACGCGCCCAAAAGCCAGCCACUCG---- ..(((((((((((...(((..((((....))))...)))((((((((....(((....)))..)))))))))).))).)))))).............---- ( -30.50) >consensus CAUUUGGGGCGUAGUUGUGCGCCGAGCUGUCGGAGUCGCUGGAUUUCGACUCGGAGUGCUGCAGAAAUCCAUCACGCACCCAAAUGGCAGCCACUCG____ ((((((((((((.((.((((.((((...(((((((((....))))))))))))).)))).)).((......)))))).))))))))............... (-35.67 = -35.95 + 0.28)

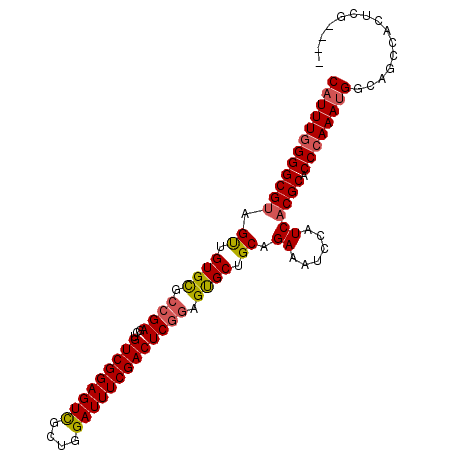

| Location | 14,587,074 – 14,587,171 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.71 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -31.73 |
| Energy contribution | -32.57 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14587074 97 - 22407834 ----CGAGUGGCUGCCAUUUGGGUGCGUGAUGGAUUUCUGCAGCACUCCGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAACUACGCCCCAAAUG ----...........((((((((.(((((.((((((((.((..........)).))))))))(((...((((....))))))).....))))))))))))) ( -35.60) >DroSec_CAF1 128799 97 - 1 ----CGAGUGGCUGCCAUUUGGGUGCGUGAUGGAUUUCUGCAGCACUCCGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAACUACGCCCCAAAUG ----...........((((((((.(((((.((((((((.((..........)).))))))))(((...((((....))))))).....))))))))))))) ( -35.60) >DroSim_CAF1 106566 97 - 1 ----CGAGUGGCUGCCAUUUGGGUGCGUGAUGGAUUUCUGCAGCACUCCGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAACUACGCCCCAAAUG ----...........((((((((.(((((.((((((((.((..........)).))))))))(((...((((....))))))).....))))))))))))) ( -35.60) >DroEre_CAF1 112422 97 - 1 ----CGAGUGGCUGCCAUUUGGGCGCGUGAUGGAUUUCUGCAGCACUCCGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAGCUACGCCCGAAAUG ----...(((((((((....))((((((((......)).))(((..((.((((((........)))))).))..)))..)))).))))))).......... ( -32.60) >DroYak_CAF1 110982 101 - 1 GAUGCGAGUGGCUGCCAUUUGGGUGCGUGAUGGAUUUCUGCAGCGCUACGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAACUACGCCCCAAAUG ...(((......)))((((((((.(((((.((((((((.((..........)).))))))))(((...((((....))))))).....))))))))))))) ( -36.30) >DroAna_CAF1 55820 97 - 1 ----CGAGUGGCUGGCUUUUGGGCGCGUGAUGGAUUUCUGCAGCACUCUGAGUCGAAAUCCAGCAACUCCGACAGCUCGGCACACAACUCCGCCCCAAAUG ----...((((((((((.(((((....((.((((((((..(((....)))....)))))))).))..))))).))).)))).)))................ ( -31.40) >consensus ____CGAGUGGCUGCCAUUUGGGUGCGUGAUGGAUUUCUGCAGCACUCCGAGUCGAAAUCCAGCGACUCCGACAGCUCGGCGCACAACUACGCCCCAAAUG ...............((((((((.(((((.((((((((.((..........)).))))))))(((...((((....))))))).....))))))))))))) (-31.73 = -32.57 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:37:03 2006