| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,584,251 – 14,584,345 |
| Length | 94 |
| Max. P | 0.946873 |

| Location | 14,584,251 – 14,584,345 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
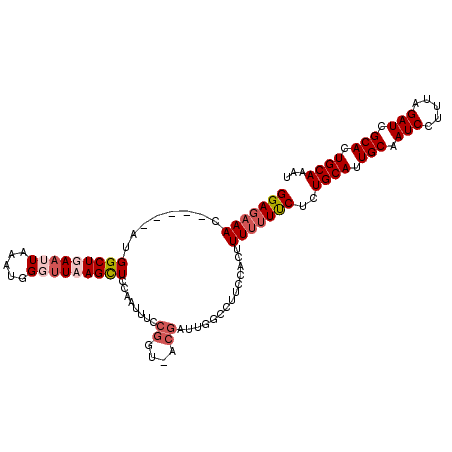
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14584251 94 + 22407834 AUUUGCAGUGCGAUCUAAAGGAUUGCAAUGCAGAGAAAAAAAGCGGAAGGU----------CGGAAAUUGGAACUGACACCAGUUAAUUCAGCCAU-----GUUUCUCC ...((((.(((((((.....))))))).))))((((((.....(....)..----------.((.......(((((....))))).......))..-----.)))))). ( -28.14) >DroSec_CAF1 125947 104 + 1 AUUUGCAGUGCGAUCUAAAGGAUUGCAAUGCAGAGAAAAAAAGUGGAAGGCCAAUCGUAACCGGAAAUGGAAGCUUAACCAAUUUAAUUCAGCCAU-----CUUUCUCC ...((((.(((((((.....))))))).))))(((((.......(((.(((...(((....)))...(((........)))..........))).)-----))))))). ( -25.41) >DroSim_CAF1 103703 104 + 1 AUUUGCACUGCGAUCUAAAGGAUUGCAAUGCAGAGAAAAAAAGUGGAAGGCCAAUCGUAACCGGAAAUGGAAGCUUAAACAAUUUAAUUCAGCCAU-----GUUUCUCC ...((((.(((((((.....))))))).))))((((((....((((...((.....))..(((....)))......................))))-----.)))))). ( -25.00) >DroYak_CAF1 107920 105 + 1 AUUUGCAGUGCGAUCUAAAGGAUUGCAGUGCAGUGGAAAAAAG-GGAAG--CCAUCGU-UCCGGAAAUUGGAACUGAACCCAGUUAAUUCAGCCAAUUCAGCUUAAUCC ..(((((.(((((((.....))))))).)))))(((((....(-(....--))....)-))))(((.((((..(((((.........)))))))))))).......... ( -31.00) >consensus AUUUGCAGUGCGAUCUAAAGGAUUGCAAUGCAGAGAAAAAAAGUGGAAGGCCAAUCGU_ACCGGAAAUGGAAACUGAAACAAGUUAAUUCAGCCAU_____CUUUCUCC .((((((.(((((((.....))))))).))))))............................((.......(((((....))))).......))............... (-19.32 = -19.32 + -0.00)

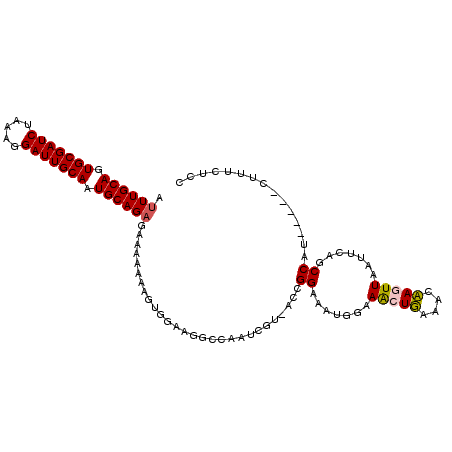
| Location | 14,584,251 – 14,584,345 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -15.29 |
| Energy contribution | -17.10 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14584251 94 - 22407834 GGAGAAAC-----AUGGCUGAAUUAACUGGUGUCAGUUCCAAUUUCCG----------ACCUUCCGCUUUUUUUCUCUGCAUUGCAAUCCUUUAGAUCGCACUGCAAAU (((((((.-----..(((.(((..(((((....)))))..........----------...))).)))...)))))))(((.(((.(((.....))).))).))).... ( -21.06) >DroSec_CAF1 125947 104 - 1 GGAGAAAG-----AUGGCUGAAUUAAAUUGGUUAAGCUUCCAUUUCCGGUUACGAUUGGCCUUCCACUUUUUUUCUCUGCAUUGCAAUCCUUUAGAUCGCACUGCAAAU ((((((((-----(.(((..(....((((((..............))))))....)..)))........)))))))))(((.(((.(((.....))).))).))).... ( -25.64) >DroSim_CAF1 103703 104 - 1 GGAGAAAC-----AUGGCUGAAUUAAAUUGUUUAAGCUUCCAUUUCCGGUUACGAUUGGCCUUCCACUUUUUUUCUCUGCAUUGCAAUCCUUUAGAUCGCAGUGCAAAU (((((...-----..((((((((......)))).))))....)))))(((((....)))))................((((((((.(((.....))).))))))))... ( -26.20) >DroYak_CAF1 107920 105 - 1 GGAUUAAGCUGAAUUGGCUGAAUUAACUGGGUUCAGUUCCAAUUUCCGGA-ACGAUGG--CUUCC-CUUUUUUCCACUGCACUGCAAUCCUUUAGAUCGCACUGCAAAU (((..(((..((((((((((((((.....)))))))..)))))))..(((-((....)--.))))-)))...)))..((((.(((.(((.....))).))).))))... ( -26.90) >consensus GGAGAAAC_____AUGGCUGAAUUAAAUGGGUUAAGCUCCAAUUUCCGGU_ACGAUUGGCCUUCCACUUUUUUUCUCUGCAUUGCAAUCCUUUAGAUCGCACUGCAAAU (((((((........(((((((((.....)))))))))........((....))..............)))))))..((((.(((.(((.....))).))).))))... (-15.29 = -17.10 + 1.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:55 2006