
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,568,636 – 14,568,732 |
| Length | 96 |
| Max. P | 0.851496 |

| Location | 14,568,636 – 14,568,732 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -12.24 |
| Energy contribution | -11.97 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
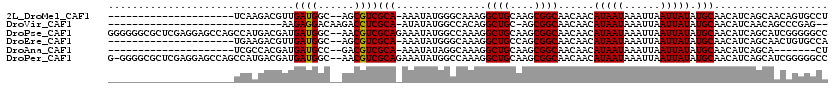
>2L_DroMel_CAF1 14568636 96 + 22407834 ---------------------UCAAGACGUUGAUGGC--AGCGUCGCA-AAAUAUGGGCAAAGGCUGCAAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCAACAGUGCCU ---------------------....(((((((....)--))))))...-......(((((...((((...(((....)...(((((......)))))..)).....)))).....))))) ( -24.70) >DroVir_CAF1 42336 87 + 1 -----------------------------AAGAGGACAAGACCUCGCA-AUAUAUGGCCACAGGCUGC-AGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAACAGCCCGAG-- -----------------------------..((((......))))((.-.......))..(.(((((.-...(....)...(((((......)))))............))))).)..-- ( -15.40) >DroPse_CAF1 113474 118 + 1 GGGGGGCGCUCGAGGAGCCAGCCAUGACGAUGAUGGC--AACGUCGCAGAAAUAUGGCCAAAGGCUGCAAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCAUCGGGGGCC ....(((.((((((.((((.((((((.(..(((((..--..)))))..)...))))))....)))).)..(((....)...(((((......)))))...........)).))))).))) ( -37.70) >DroEre_CAF1 95012 96 + 1 ---------------------UGAAGACGUUGAUGGC--AGCGUCGCA-AAAUAUGGGCAAAGGCUGCCAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCAACUGUGCCA ---------------------....(((((((....)--))))))...-.......((((.((((((....)))).......................(((.......))).)).)))). ( -25.00) >DroAna_CAF1 38100 89 + 1 ---------------------UCGCCACGAUGAUGCC--GACGUCGCA-AAAUAUAGGCAAAGGCUGCAAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCA-------CU ---------------------..((...((((.(((.--...(((((.-.......(((....)))....)))))......(((((......)))))..))).)))).)).-------.. ( -17.70) >DroPer_CAF1 117807 117 + 1 G-GGGGCGCUCGAGGAGCCAGCCAUGACGAUGAUGGC--AACGUCGCAGAAAUAUGGCCAAAGGCUGCAAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCAUCGGGGGCC .-..(((.((((((.((((.((((((.(..(((((..--..)))))..)...))))))....)))).)..(((....)...(((((......)))))...........)).))))).))) ( -37.70) >consensus _____________________UCAAGACGAUGAUGGC__AACGUCGCA_AAAUAUGGCCAAAGGCUGCAAGCGGCAACAACAUAAUAAAUUAAUUAUAUGCAACAUCAGCAACGGGGGCC ...............................((((......))))(((...............((((....))))......(((((......))))).)))................... (-12.24 = -11.97 + -0.28)


| Location | 14,568,636 – 14,568,732 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -15.15 |
| Energy contribution | -15.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
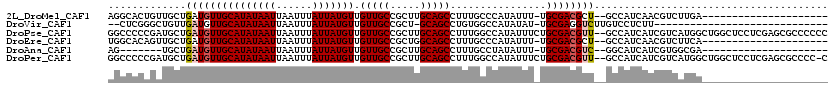
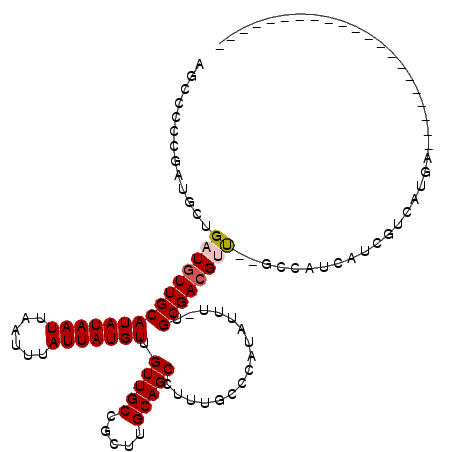
>2L_DroMel_CAF1 14568636 96 - 22407834 AGGCACUGUUGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUUGCAGCCUUUGCCCAUAUUU-UGCGACGCU--GCCAUCAACGUCUUGA--------------------- .((((.((((((.(((((.(((((((((......))))))).(((((.....)))))....))..))))).-.)))))).)--))).............--------------------- ( -25.70) >DroVir_CAF1 42336 87 - 1 --CUCGGGCUGUUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCU-GCAGCCUGUGGCCAUAUAU-UGCGAGGUCUUGUCCUCUU----------------------------- --(.((((((((.(..((.(((((((((......)))))).))).))..).-)))))))).)((.......-.))((((......))))..----------------------------- ( -25.90) >DroPse_CAF1 113474 118 - 1 GGCCCCCGAUGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUUGCAGCCUUUGGCCAUAUUUCUGCGACGUU--GCCAUCAUCGUCAUGGCUGGCUCCUCGAGCGCCCCCC (((.(.(((.((((((((((((((((((......))))))).(((((.....)))))................))))))))--(((((.......))))).)))...))).).))).... ( -32.00) >DroEre_CAF1 95012 96 - 1 UGGCACAGUUGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUGGCAGCCUUUGCCCAUAUUU-UGCGACGCU--GCCAUCAACGUCUUCA--------------------- (((((..(((((.(((((.(((((((((......))))))).((((((...))))))....))..))))).-.)))))..)--))))............--------------------- ( -27.00) >DroAna_CAF1 38100 89 - 1 AG-------UGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUUGCAGCCUUUGCCUAUAUUU-UGCGACGUC--GGCAUCAUCGUGGCGA--------------------- .(-------(((((((((((((((((((......))))))).(((((.....)))))..............-.))))))))--)))))...........--------------------- ( -27.60) >DroPer_CAF1 117807 117 - 1 GGCCCCCGAUGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUUGCAGCCUUUGGCCAUAUUUCUGCGACGUU--GCCAUCAUCGUCAUGGCUGGCUCCUCGAGCGCCCC-C (((.(.(((.((((((((((((((((((......))))))).(((((.....)))))................))))))))--(((((.......))))).)))...))).).)))..-. ( -32.00) >consensus AGCCCCCGAUGCUGAUGUUGCAUAUAAUUAAUUUAUUAUGUUGUUGCCGCUUGCAGCCUUUGCCCAUAUUU_UGCGACGUU__GCCAUCAUCGUCAUGA_____________________ .............(((((((((((((((......))))))).(((((.....)))))................))))))))....................................... (-15.15 = -15.43 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:48 2006