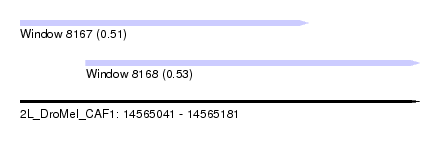
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,565,041 – 14,565,181 |
| Length | 140 |
| Max. P | 0.526260 |

| Location | 14,565,041 – 14,565,142 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -16.61 |
| Energy contribution | -18.17 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
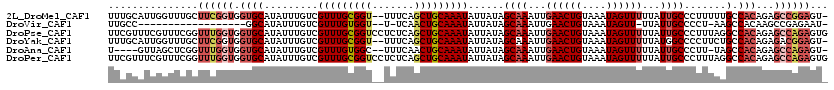
>2L_DroMel_CAF1 14565041 101 + 22407834 ---UCUGUCUGUGGUUGUCGGUUUGGUUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGUUUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGU ---...............(((((..((((((....(((..(....)..).)).........(((((((((.....)))))))))......))))))..))))). ( -30.40) >DroVir_CAF1 38038 85 + 1 CUGCCAGUCUGCGGUUGCUGCUUUUGUUGCC------------------GGCAUAUUUGUCGUUUGUGGUU-UCAACUGCAAAUAUUAUAGCAAAUUGAACUGU ....((((.((((((.((.......)).)))------------------((((....))))(((((..((.-...))..)))))......))).))))...... ( -21.60) >DroSim_CAF1 84130 101 + 1 ---UCUGUCUGUGGUUGUCGGUUUGGUUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGUUUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGU ---...............(((((..((((((....(((..(....)..).)).........(((((((((.....)))))))))......))))))..))))). ( -30.40) >DroEre_CAF1 90764 99 + 1 ----CUGUCUGUGGCUGUCGGUUUGGUUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGUUUUC-GCUGCAAAUAUUAUAGCAAAUUGAACUGU ----..............(((((..((((((....(((..(....)..).)).........(((((((((....-)))))))))......))))))..))))). ( -29.90) >DroYak_CAF1 88604 101 + 1 ---UCUGUCUGUGGUUGUCGGUUUGGUUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGUUUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGU ---...............(((((..((((((....(((..(....)..).)).........(((((((((.....)))))))))......))))))..))))). ( -30.40) >DroAna_CAF1 34896 95 + 1 ---CCUGUCUGUGGCUGU--GUUUGUU----GUUAGCUCGGUUUGGUGGUGCAUAUUUGUCGUUUGUGGCUUUCAACUGCAAAUAUUAUAGCAAAUUGAACUGU ---...((..(..((...--....)).----.)..)).(((((..((..(((((((((((.(((.(......).))).))))))))....))).))..))))). ( -20.30) >consensus ___UCUGUCUGUGGUUGUCGGUUUGGUUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGUUUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGU ..................(((((((((((((...................(((....))).(((((((((.....)))))))))......))))))))))))). (-16.61 = -18.17 + 1.56)

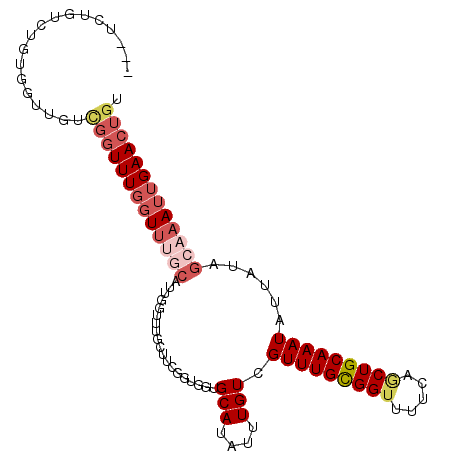
| Location | 14,565,064 – 14,565,181 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.92 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -19.35 |
| Energy contribution | -20.68 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14565064 117 + 22407834 UUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGU--UUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUUGCCCUUUUUGCCACAGAGCCGGAGU- .............((((((((.(((((........(((((((((--.....)))))))))......((((...((((((....))))))...))))......)).)))...))))))))- ( -32.90) >DroVir_CAF1 38064 96 + 1 UUGCC------------------GGCAUAUUUGUCGUUUGUGGU--U-UCAACUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUU-UUAUUGCCCCU-AAGCCACAAGCCGAGAAU- .....------------------((((....))))(((((((((--(-(.....((((..(((((.(((........)))..))))).-...))))....-))))))))))).......- ( -21.50) >DroPse_CAF1 108824 120 + 1 UUCGUUUCGUUUCGGUUUGGUGGUGCAUAUUUGUCGUUUGCGGUCCUCUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUUGCCCUUUAGGCCACAGAGCCAGAGUG .............(((((.(((((...........(((((((((.......)))))))))......((((...((((((....))))))...)))).......))))).)))))...... ( -31.10) >DroYak_CAF1 88627 117 + 1 UUUGCAUUGGUUUGCUUCGGUGGUGCAUAUUUGUCGUUUGCGGU--UUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUGGCCCCUUCUGCCACAGAGACGGAGU- (((((((..(((((((..((..(.......)..))(((((((((--.....))))))))).....)))))))..)..))))))..((((((.((((.......)))).)))))).....- ( -33.90) >DroAna_CAF1 34917 112 + 1 U----GUUAGCUCGGUUUGGUGGUGCAUAUUUGUCGUUUGUGGC--UUUCAACUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUUGCCCUU-UAGCCACAGAGCCAGAGU- .----....(((((((((.(((((..((((((((.(((.(....--..).))).))))))))....((((...((((((....))))))...))))....-..))))).))))).))))- ( -30.70) >DroPer_CAF1 113101 120 + 1 UUCGUUUCGUUUCGGUUUGGUGGUGCAUAUUUGUCGUUUGCGGUCCUCUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUUGCCCUUUAGGCCACAGAGCCAGAGUG .............(((((.(((((...........(((((((((.......)))))))))......((((...((((((....))))))...)))).......))))).)))))...... ( -31.10) >consensus UUCGCAUUGGUUCGGUUUGGUGGUGCAUAUUUGUCGUUUGCGGU__UUUCAGCUGCAAAUAUUAUAGCAAAUUGAACUGUAAAUAGUUUUUAUUGCCCUUUAAGCCACAGAGCCAGAGU_ ...............((((((.((((.........(((((((((.......)))))))))......((((...((((((....))))))...)))).......).)))...))))))... (-19.35 = -20.68 + 1.33)
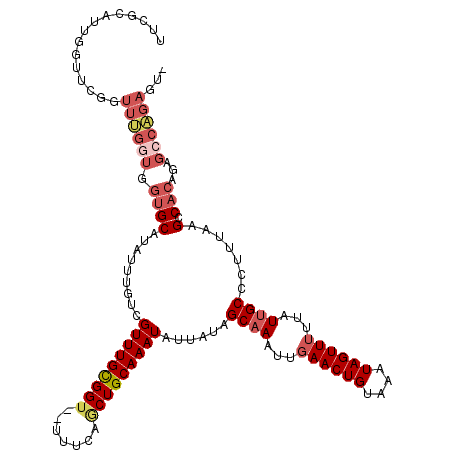
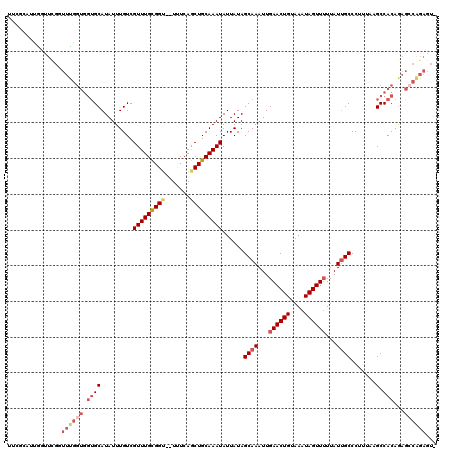
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:44 2006