
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,560,808 – 14,560,908 |
| Length | 100 |
| Max. P | 0.915437 |

| Location | 14,560,808 – 14,560,908 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -14.95 |
| Energy contribution | -16.65 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.710958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
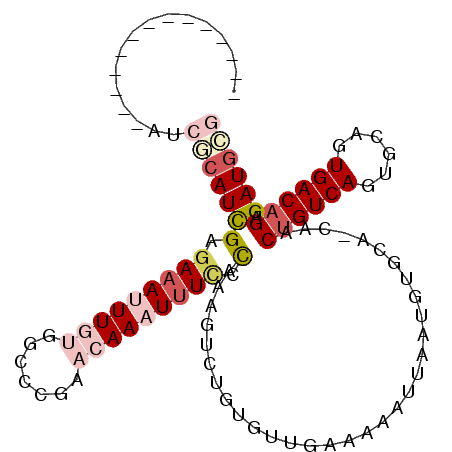
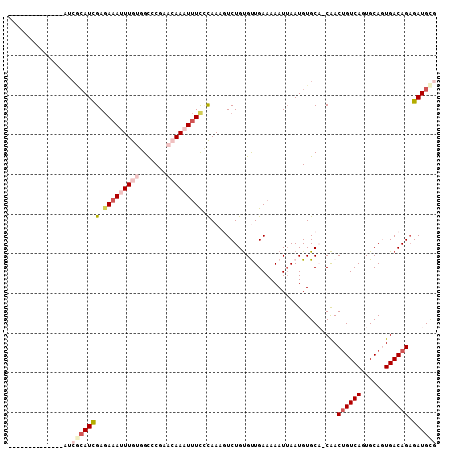
>2L_DroMel_CAF1 14560808 100 + 22407834 ------GCAUC-GCAUCGCAUCGAGAAAUUUGUGGCCCGAACAAAUUUCCCAAAGUCUGUGUUGAAAAAUUAAUGUGCA-CAACUGUCAGUGCAGUGACAGAGAUGCG ------.....-....(((((((.(((((((((.......))))))))).)....((((((((((....))))).((((-(........)))))...))))))))))) ( -28.60) >DroVir_CAF1 34383 89 + 1 --------------GACACAUCGAAAAAUUUGUUGCUCC---AAUUCUC-UAAACUCUGUGCUGAAAAAUUAAUGUGCC-CAACUGUCAUUGCAAUGACAGAGAUGAA --------------....((((.................---.......-........(..(............)..).-...(((((((....))))))).)))).. ( -13.80) >DroSim_CAF1 79895 93 + 1 --------------AUCGCAUCGAGAAAUUUGUGGCCCGAACAAAUUUCCCAAAGUCUGUGUUGAAAAAUUAAUGUGCA-CAACUGUCAGUGCAGUGACAGAGAUGCG --------------..(((((((.(((((((((.......))))))))).)....((((((((((....))))).((((-(........)))))...))))))))))) ( -28.60) >DroYak_CAF1 83884 107 + 1 GCAUCAGCAUCAGCAUCGCAUCGAGAAAUUUGUGGCCCGAACAAAUUUCCCAAAGUCUGUGUUGAAAAAUUAAUGUGCA-CAACUGUCAGUGCAGUGACAGAGAUGCG (((((.((((((((((.((...(.(((((((((.......))))))))).)...))..)))))))..........))).-...((((((......)))))).))))). ( -31.30) >DroMoj_CAF1 41231 90 + 1 ---------------UCACAUUGAGAAAUUUGUUGUUCC---AAUUUUCGUAAAUUCUGUGCUGAAAAAUGAAUGUGCCCCAACAGUCAUUGCAUUGACAGCGAUGUG ---------------.(((((((.((((.(((......)---)).)))).......((((.(.....(((((.(((......))).))))).....)))))))))))) ( -19.70) >DroAna_CAF1 31181 88 + 1 -------------------AUCGAGAAAUUUGUGGCCCGAACAAAUUUUCCAAAGUCUGCGUUGAAAAAUUAAUGUGCA-CGACUGUCACUGCAGUGACAGAGAUGCG -------------------...(..((((((((.......))))))))..).......(((((((....)))))))(((-...(((((((....)))))))...))). ( -24.80) >consensus ______________AUCGCAUCGAGAAAUUUGUGGCCCGAACAAAUUUCCCAAAGUCUGUGUUGAAAAAUUAAUGUGCA_CAACUGUCAGUGCAGUGACAGAGAUGCG ................(((((((.(((((((((.......))))))))).)................................((((((......)))))).)))))) (-14.95 = -16.65 + 1.70)

| Location | 14,560,808 – 14,560,908 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.65 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -15.27 |
| Energy contribution | -17.58 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
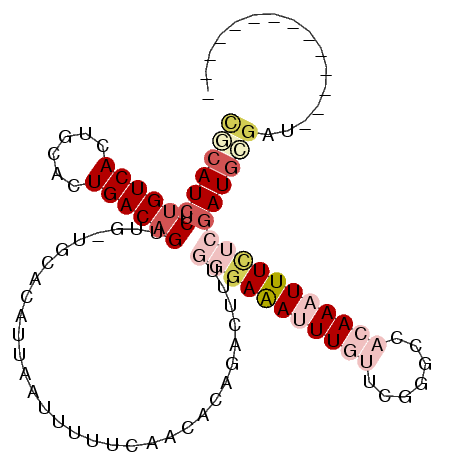
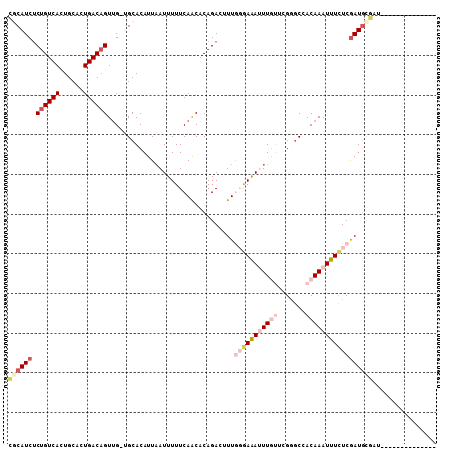
>2L_DroMel_CAF1 14560808 100 - 22407834 CGCAUCUCUGUCACUGCACUGACAGUUG-UGCACAUUAAUUUUUCAACACAGACUUUGGGAAAUUUGUUCGGGCCACAAAUUUCUCGAUGCGAUGC-GAUGC------ ((((((.((((((......))))))...-.(((..........((......))..(((((((((((((.......)))))))))))))))))))))-)....------ ( -29.60) >DroVir_CAF1 34383 89 - 1 UUCAUCUCUGUCAUUGCAAUGACAGUUG-GGCACAUUAAUUUUUCAGCACAGAGUUUA-GAGAAUU---GGAGCAACAAAUUUUUCGAUGUGUC-------------- ..((...(((((((....))))))).))-((((((((.....(((......)))....-(((((((---(......)))..)))))))))))))-------------- ( -19.60) >DroSim_CAF1 79895 93 - 1 CGCAUCUCUGUCACUGCACUGACAGUUG-UGCACAUUAAUUUUUCAACACAGACUUUGGGAAAUUUGUUCGGGCCACAAAUUUCUCGAUGCGAU-------------- ((((((.((((((......))))))(((-((................))))).....(((((((((((.......)))))))))))))))))..-------------- ( -27.89) >DroYak_CAF1 83884 107 - 1 CGCAUCUCUGUCACUGCACUGACAGUUG-UGCACAUUAAUUUUUCAACACAGACUUUGGGAAAUUUGUUCGGGCCACAAAUUUCUCGAUGCGAUGCUGAUGCUGAUGC .((((..((((((......))))))..)-))).(((((.....(((.((..(...(((((((((((((.......)))))))))))))..)..)).)))...))))). ( -31.60) >DroMoj_CAF1 41231 90 - 1 CACAUCGCUGUCAAUGCAAUGACUGUUGGGGCACAUUCAUUUUUCAGCACAGAAUUUACGAAAAUU---GGAACAACAAAUUUCUCAAUGUGA--------------- (((((.((((..((((.((((..(((....)))))))))))...))))...........((((.((---(......))).))))...))))).--------------- ( -18.30) >DroAna_CAF1 31181 88 - 1 CGCAUCUCUGUCACUGCAGUGACAGUCG-UGCACAUUAAUUUUUCAACGCAGACUUUGGAAAAUUUGUUCGGGCCACAAAUUUCUCGAU------------------- .((((..(((((((....)))))))..)-)))(((..((((((((((........)))))))))))))(((((..........))))).------------------- ( -25.20) >consensus CGCAUCUCUGUCACUGCACUGACAGUUG_UGCACAUUAAUUUUUCAACACAGACUUUGGGAAAUUUGUUCGGGCCACAAAUUUCUCGAUGCGAU______________ ((((((.((((((......))))))................................(((((((((((.......)))))))))))))))))................ (-15.27 = -17.58 + 2.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:40 2006