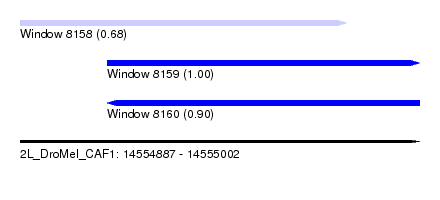
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,554,887 – 14,555,002 |
| Length | 115 |
| Max. P | 0.999871 |

| Location | 14,554,887 – 14,554,981 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -17.71 |
| Energy contribution | -17.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
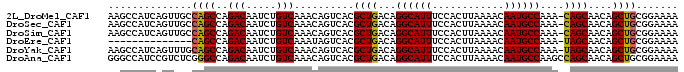
>2L_DroMel_CAF1 14554887 94 + 22407834 AAGCCAUCAGUUGCCAGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAA ...((..((((((.......(((.....)))..........((((...((((((............))))))...-))))..)))))).)).... ( -22.60) >DroSec_CAF1 86662 94 + 1 AAGCCAUCAGUUGCCAGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAA ...((..((((((.......(((.....)))..........((((...((((((............))))))...-))))..)))))).)).... ( -22.60) >DroSim_CAF1 74128 94 + 1 AAGCCAUCAGUUGCCAGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAA ...((..((((((.......(((.....)))..........((((...((((((............))))))...-))))..)))))).)).... ( -22.60) >DroEre_CAF1 81036 80 + 1 --------------CAGCCAGACAAUCUGUCAAAUAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-UAGCAACAGCUGCGGAAAA --------------((((..(((.....)))..........((((...((((((............))))))...-))))....))))....... ( -17.10) >DroYak_CAF1 77951 94 + 1 AAGCCAUCAGUUUGCAGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-UAGCAACAGCUGCGGAAAA ..........((((((((..(((.....)))..........((((...((((((............))))))...-))))....))))))))... ( -23.80) >DroAna_CAF1 25307 95 + 1 GGGCCAUCCGUCUCGGGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAGCCAGCAACAGCUGCGGAAAA ......(((((....((((((.....(((.....))).....)))...((((((............))))))..)))(((....))))))))... ( -24.60) >consensus AAGCCAUCAGUUGCCAGCCAGACAAUCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA_CAGCAACAGCUGCGGAAAA ..............((((..(((.....)))..........((((...((((((............))))))....))))....))))....... (-17.71 = -17.52 + -0.19)
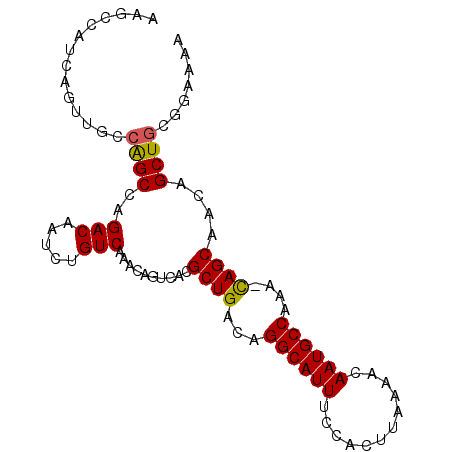
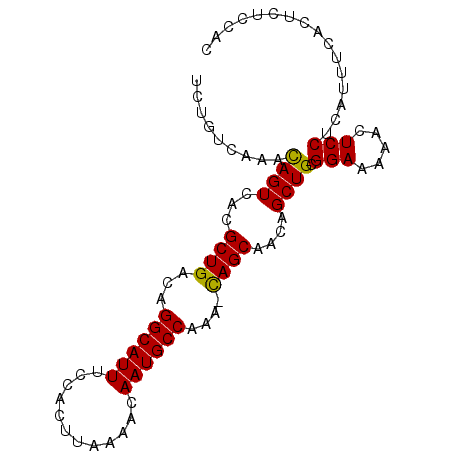
| Location | 14,554,912 – 14,555,002 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 97.34 |
| Mean single sequence MFE | -15.50 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.75 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.32 |
| SVM RNA-class probability | 0.999871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14554912 90 + 22407834 UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUCCAC .........((((...((((...((((((............))))))...-))))....)))).(((.....)))................ ( -16.10) >DroSec_CAF1 86687 90 + 1 UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUCCAC .........((((...((((...((((((............))))))...-))))....)))).(((.....)))................ ( -16.10) >DroSim_CAF1 74153 90 + 1 UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-CAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUCCAC .........((((...((((...((((((............))))))...-))))....)))).(((.....)))................ ( -16.10) >DroEre_CAF1 81047 89 + 1 UCUGUCAAAUAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-UAGCAACAGCUGCGGAAAAACUCCUCAU-UCACUCUUCAC (((((......(((.....))).((((((............))))))...-.(((....))))))))............-........... ( -14.10) >DroYak_CAF1 77976 90 + 1 UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA-UAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUUCAC .........((((...((((...((((((............))))))...-))))....)))).(((.....)))................ ( -14.30) >DroAna_CAF1 25332 91 + 1 UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAGCCAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUCCAC ..(((((...........)))))((((((............))))))..((.(((....)))))(((.....)))................ ( -16.30) >consensus UCUGUCAAACAGUCACGCUGACAGGCAUUUCCACUUAAAACAAUGCCAAA_CAGCAACAGCUGCGGAAAAACUCCUCAUUUCACUCUCCAC .........((((...((((...((((((............))))))....))))....)))).(((.....)))................ (-15.11 = -14.75 + -0.36)

| Location | 14,554,912 – 14,555,002 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.34 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -22.95 |
| Energy contribution | -22.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14554912 90 - 22407834 GUGGAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUG-UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA ((((((((.............)))))))).......((((-...((((((............))))))...))))....(((.....))). ( -24.42) >DroSec_CAF1 86687 90 - 1 GUGGAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUG-UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA ((((((((.............)))))))).......((((-...((((((............))))))...))))....(((.....))). ( -24.42) >DroSim_CAF1 74153 90 - 1 GUGGAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUG-UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA ((((((((.............)))))))).......((((-...((((((............))))))...))))....(((.....))). ( -24.42) >DroEre_CAF1 81047 89 - 1 GUGAAGAGUGA-AUGAGGAGUUUUUCCGCAGCUGUUGCUA-UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUAUUUGACAGA ((.((((((.(-.(((.(.((.((((((((((...((((.-...))))..)))....))))))).)).).)))...).))).))).))... ( -22.00) >DroYak_CAF1 77976 90 - 1 GUGAAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUA-UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA (((((.(((.(..(((.(.((.((((((((((...((((.-...))))..)))....))))))).)).).)))...).))).))).))... ( -22.80) >DroAna_CAF1 25332 91 - 1 GUGGAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUGGCUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA ((((((((.............)))))))).......((((((..((((((............)))))).))))))....(((.....))). ( -27.82) >consensus GUGGAGAGUGAAAUGAGGAGUUUUUCCGCAGCUGUUGCUG_UUUGGCAUUGUUUUAAGUGGAAAUGCCUGUCAGCGUGACUGUUUGACAGA ((.((((((.(..(((.(.((.((((((((((...((((.....))))..)))....))))))).)).).)))...).))).))).))... (-22.95 = -22.73 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:37 2006