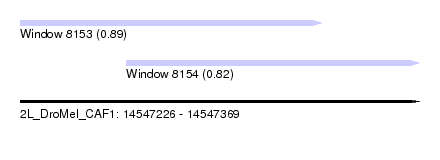
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,547,226 – 14,547,369 |
| Length | 143 |
| Max. P | 0.894399 |

| Location | 14,547,226 – 14,547,334 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.92 |
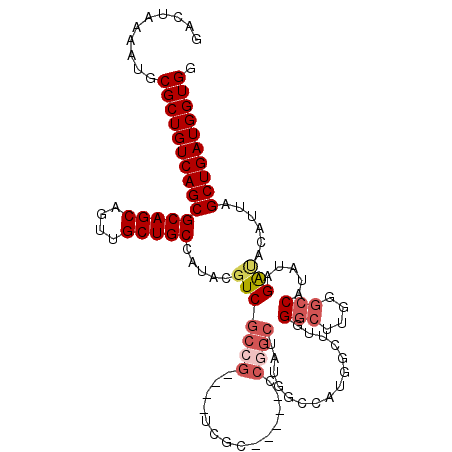
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.14 |
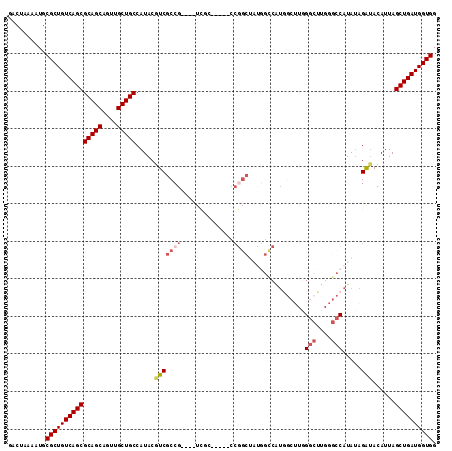
| Combinations/Pair | 1.22 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
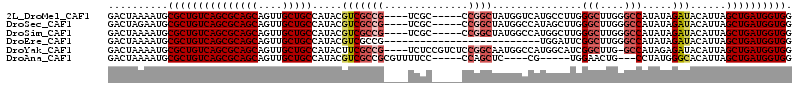
>2L_DroMel_CAF1 14547226 108 + 22407834 CACGACAGGGCAA--GAUUUAUUUUCGGUAUUCCAUGGGCGACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG----UCGCCCGGCUAUGGU-CAUGCCUUG .....(((((((.--((((.((..((((....))..(((((((......(((.(((..(.(((((....))))))..))).))).)----))))))))..)))))-).))))))) ( -40.80) >DroVir_CAF1 20382 100 + 1 CACGACAUGCCAA--GAUUUAUUUUUGGUAUUCCACGAGCGACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGACCCA--UCGUUCCG------UCCUCU----- ..(((((((((((--((.....)))))))))......(((((((....(((((.....)))))..))))))).......)))).....--........------......----- ( -31.40) >DroEre_CAF1 73649 91 + 1 CACGACAGGGCAA--GAUUUAUUUUCGGCCUUCCAUGGGCGACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG----------------------UGGAUUC .......((((..--((......))..))))(((((((.((((.....(((((.....)))))((((...)))).....)))))))----------------------))))... ( -33.70) >DroMoj_CAF1 24915 100 + 1 CACGACAUGCCAA--GAUUUAUUUUCGGUAUUCCACGAGCAACCCAAAUGCGCUGUCAGCACAGCAGUUGCUGCCAUACGUCGUCCCA--UCGCUCGG------UCCUCU----- ...((((((((.(--((.....))).)))))....(((((........(((.......)))((((....))))...............--..))))))------))....----- ( -23.20) >DroAna_CAF1 17835 103 + 1 CAUGACAUGAAAA--GAUUUAUUUUCGGUAUUCCAUGGGCGACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCGCGUUUUCCCCAGCUC-----CG-----UG ((((.((((((((--......)))))((....)))))((((((.....(((((.....)))))((((...)))).....))))))((..........))..-----))-----)) ( -29.70) >DroPer_CAF1 93109 106 + 1 CACGAGAUGGCAAGAGAUUUAUUUUCGGUAUUCCAUGGGCGACUAAAAUGCGCUGUCAGCGCACCAGUUGCCGCCAUACGUCGCCC----CAGCCAGGCACAGAUCCACA----- .(((..(((((..((((.....))))((....))...(((((((....(((((.....)))))..)))))))))))).))).(((.----......)))...........----- ( -34.10) >consensus CACGACAUGGCAA__GAUUUAUUUUCGGUAUUCCAUGGGCGACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCC____UCGCCCGG______U_CACG_____ ..((((....................((....))...(((((((....(((((.....)))))..))))))).......))))................................ (-21.62 = -21.48 + -0.14)



| Location | 14,547,264 – 14,547,369 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.77 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14547264 105 + 22407834 GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG----UCGC-----CCGGCUAUGGUCAUGCCUUGGGCUUGGGCCAUAUAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((----....-----.))))(((((((..(((...)))...)))))))..)))......)))))))))). ( -44.30) >DroSec_CAF1 79118 105 + 1 GACUAGAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG----UCGC-----CCGGCUAUGGCCAUAGCUUGGGCUUGGGCCAUAUAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((----....-----.))))(((((((..(((....)))..)))))))..)))......)))))))))). ( -46.20) >DroSim_CAF1 64926 105 + 1 GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG----UCGC-----CCGGCUAUGGCCAUGGCUUGGGCUUGGGCCAUAUAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((----....-----.))))(((((((..(((....)))..)))))))..)))......)))))))))). ( -46.20) >DroEre_CAF1 73687 88 + 1 GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG--------------------------UGGAUUCGGCUUGGGCCAUAUAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((--------------------------......))))...)))...............)))))))))). ( -31.30) >DroYak_CAF1 70033 109 + 1 GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACUUCGCCG----UCUCCGUCUCCGGCAAUGGCCAUGGCAUCGGCUUG-GCCAUAGAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((----..........)))).(((((((.(((....)))))-))))).)))........)))))))))). ( -44.40) >DroAna_CAF1 17873 97 + 1 GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCGCGUUUUCC-----CCAGCUC----CG-----UGGAACUG---CCUAUGGGCACAUUAGCUGAUGGUGG ..........(((((((((((((((....)))))...((((....)))).....-----...((.(----((-----(((.....---.))))))))......)))))))))). ( -35.30) >consensus GACUAAAAUGCGCUGUCAGCGCAGCAGUUGCUGCCAUACGUCGCCG____UCGC_____CCGGCUAUGGCCAUGGCUUGGGCUUGGGCCAUAUAGAUACAUUAGCUGAUGGUGG ..........(((((((((((((((....))))).....(((((((..............))))...............(((....))).....)))......)))))))))). (-28.69 = -29.77 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:31 2006