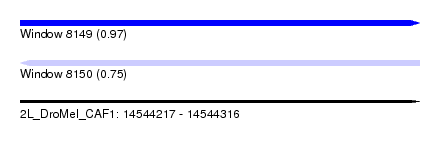
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 14,544,217 – 14,544,316 |
| Length | 99 |
| Max. P | 0.966545 |

| Location | 14,544,217 – 14,544,316 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -18.68 |
| Energy contribution | -20.00 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 14544217 99 + 22407834 GCACCCGCUUCGAGCGCAAAGCCCAAAGCCCGAAGCCCCUUUUGGCCCCGGUAGAAUUCUACCCCAAAUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ......((((((.((............)).))))))....(((((....(((((....))))))))))....(((((.(((((....))))).))))). ( -29.20) >DroSec_CAF1 76075 98 + 1 GCACCCGCUUCGAGCGCAAAGCCCAAAGCCCGAAGCCCCU-UUGGCCCCGGUAGAAUUCUACCCCAAAUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ......((((((.((............)).))))))...(-((((....(((((....))))))))))....(((((.(((((....))))).))))). ( -29.10) >DroSim_CAF1 61887 98 + 1 GCACCCGCUUCGAGCGCAAAGCCCAAAGCCCGAAGCCCCU-UUGGCCCCUGUAGAAUUCUACCCCAAAUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ......((((((.((............)).))))))...(-((((.....((((....)))).)))))....(((((.(((((....))))).))))). ( -25.40) >DroEre_CAF1 70986 86 + 1 GCACCCGCCUCGAGCGCAAAGCCCAAAGCCCAAGGCCC----------A--AACAAUUCUACCCCA-AUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ......((((.(.((............)).).))))..----------.--...............-.....(((((.(((((....))))).))))). ( -16.40) >DroYak_CAF1 67017 88 + 1 GCACCCGCUUCUAGUGCAAAGCCCAAAGCCCAAAGCCC----------CGGUACCAUUCUACCCCA-GUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ((((.........)))).....................----------.((((......))))...-.....(((((.(((((....))))).))))). ( -15.70) >consensus GCACCCGCUUCGAGCGCAAAGCCCAAAGCCCGAAGCCCCU_UUGGCCCCGGUAGAAUUCUACCCCAAAUUCAGCAGAAAAGAUAUAUAUUUUGUCUGCA ......((((((.((............)).)))))).............(((((....))))).........(((((.(((((....))))).))))). (-18.68 = -20.00 + 1.32)


| Location | 14,544,217 – 14,544,316 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -24.66 |
| Energy contribution | -27.58 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
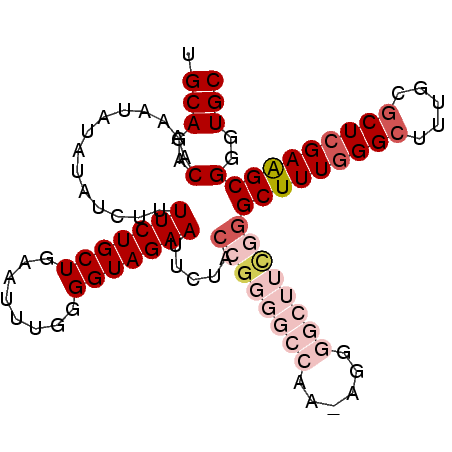
>2L_DroMel_CAF1 14544217 99 - 22407834 UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAUUUGGGGUAGAAUUCUACCGGGGCCAAAAGGGGCUUCGGGCUUUGGGCUUUGCGCUCGAAGCGGGUGC .(((..(..............(((((((........))))))).....((((((((......))))))))(((((((((.....))))))))))..))) ( -37.00) >DroSec_CAF1 76075 98 - 1 UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAUUUGGGGUAGAAUUCUACCGGGGCCAA-AGGGGCUUCGGGCUUUGGGCUUUGCGCUCGAAGCGGGUGC .(((..(..............(((((((........))))))).....((((((((..-...))))))))(((((((((.....))))))))))..))) ( -36.80) >DroSim_CAF1 61887 98 - 1 UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAUUUGGGGUAGAAUUCUACAGGGGCCAA-AGGGGCUUCGGGCUUUGGGCUUUGCGCUCGAAGCGGGUGC .(((..(..............(((((((........))))))).......((((((..-...))))))..(((((((((.....))))))))))..))) ( -31.20) >DroEre_CAF1 70986 86 - 1 UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAU-UGGGGUAGAAUUGUU--U----------GGGCCUUGGGCUUUGGGCUUUGCGCUCGAGGCGGGUGC ..(((((((..(((...((..(((....))).-.)).)))...)))))--)----------).((((...(((((((((.....))))))))))))).. ( -26.40) >DroYak_CAF1 67017 88 - 1 UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAC-UGGGGUAGAAUGGUACCG----------GGGCUUUGGGCUUUGGGCUUUGCACUAGAAGCGGGUGC .(((..(........((((..(((((((....-...))))))).))))(((----------((((.....)))))))(((((......))))))..))) ( -25.00) >consensus UGCAGACAAAAUAUAUAUCUUUUCUGCUGAAUUUGGGGUAGAAUUCUACCGGGGCCAA_AGGGGCUUCGGGCUUUGGGCUUUGCGCUCGAAGCGGGUGC .(((..(..............(((((((........))))))).....((((((((......))))))))(((((((((.....))))))))))..))) (-24.66 = -27.58 + 2.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:28 2006